
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,516,582 – 5,516,681 |
| Length | 99 |
| Max. P | 0.990240 |

| Location | 5,516,582 – 5,516,681 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.08 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.55 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
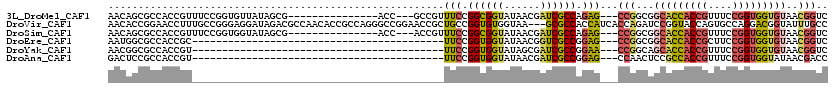
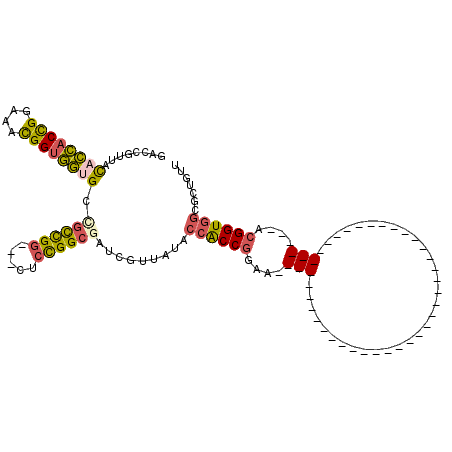
>3L_DroMel_CAF1 5516582 99 + 23771897 AACAGCGCCACCGUUUCCGGUGUUAUAGCG---------------ACC---GCCGUUUCCGGCGGUAUAACGAUCGCCAGAG---CCGGCGGCACCACCGUUUCCGGUGGUGUAACGGUC ....((..(((((....))))).....))(---------------(((---((((.(((.((((((......)))))).)))---.)))).(((((((((....)))))))))...)))) ( -49.60) >DroVir_CAF1 62020 117 + 1 AACACCGGAACCUUUGCCGGGAGGAUAGACGCCAACACCGCCAGGGCCGGAACCGCUGCCGGUGUGGUAA---GCGCCACCAUCACCAGAUCCGGUACCAGUGCCAGGACGGUAUUUGCC ....((((........))))..........((.((.((((((..((((((.((((...(.((((((((..---.....)))).)))).)...)))).)).).))).)).)))).)).)). ( -39.90) >DroSim_CAF1 79803 99 + 1 AACAGCGCCACCGUUUCCGGUGGUAUAGCG---------------ACC---ACCGUUUCCGGCGGUAUAACGAUCGCCAGAG---CCGGCGGCACCACCGUUUCCGGUGGUGUAACGGUC ....(((((((((....)))))))...))(---------------(((---.(((.(((.((((((......)))))).)))---.)))..(((((((((....)))))))))...)))) ( -49.80) >DroEre_CAF1 79163 75 + 1 AAUGGCGCCACCGC------------------------------------------UUCCGGUGGUAUAACGGUCGCCGGAG---CCGGCGGCACCACCGCUUCCGGUGGUGUAACGGUC ....(((((((((.------------------------------------------...(((((((......(((((((...---.))))))))))))))....)))))))))....... ( -41.40) >DroYak_CAF1 67259 75 + 1 AACGGCGCCACCGU------------------------------------------UUCCGGUGGUAUAGCGAUCGCCGGAA---CCGGCAGCACCACCGUUUCCGGUGGUGUAACGGUC ....(((((((((.------------------------------------------...)))))))...))((((((((...---.)))).(((((((((....)))))))))...)))) ( -40.00) >DroAna_CAF1 40584 75 + 1 GACUCCGCCACCGU------------------------------------------UUCCGGUGGUAUAACGAUCGCCGGAG---CCAACUCCGCCACCGUUUCCGGUGGUAUAACGACC ......(((((((.------------------------------------------...))))))).......(((..((((---....))))(((((((....)))))))....))).. ( -30.10) >consensus AACAGCGCCACCGU__________________________________________UUCCGGUGGUAUAACGAUCGCCAGAG___CCGGCGGCACCACCGUUUCCGGUGGUGUAACGGUC ........................................................(((.((((((......)))))).)))...(((...(((((((((....)))))))))..))).. (-21.54 = -21.55 + 0.01)
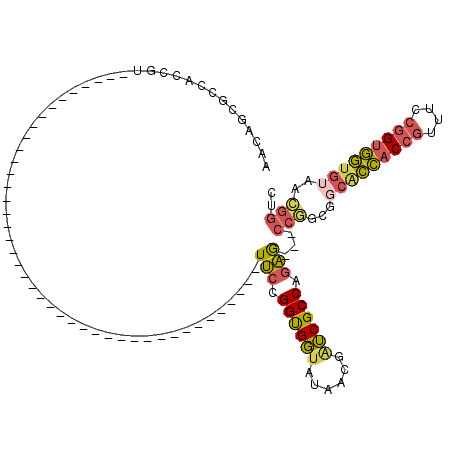
| Location | 5,516,582 – 5,516,681 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.08 |
| Mean single sequence MFE | -49.62 |
| Consensus MFE | -26.36 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.43 |
| Mean z-score | -4.70 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
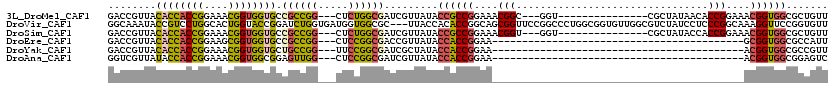
>3L_DroMel_CAF1 5516582 99 - 23771897 GACCGUUACACCACCGGAAACGGUGGUGCCGCCGG---CUCUGGCGAUCGUUAUACCGCCGGAAACGGC---GGU---------------CGCUAUAACACCGGAAACGGUGGCGCUGUU ((((....((((((((....))))))))..((((.---.(((((((..........)))))))..))))---)))---------------)((.....(((((....)))))..)).... ( -58.50) >DroVir_CAF1 62020 117 - 1 GGCAAAUACCGUCCUGGCACUGGUACCGGAUCUGGUGAUGGUGGCGC---UUACCACACCGGCAGCGGUUCCGGCCCUGGCGGUGUUGGCGUCUAUCCUCCCGGCAAAGGUUCCGGUGUU ((......)).....((((((((.(((....((((.((.((((((((---(....((((((.(((.(((....)))))).)))))).)))).))))).))))))....))).)))))))) ( -53.60) >DroSim_CAF1 79803 99 - 1 GACCGUUACACCACCGGAAACGGUGGUGCCGCCGG---CUCUGGCGAUCGUUAUACCGCCGGAAACGGU---GGU---------------CGCUAUACCACCGGAAACGGUGGCGCUGUU ((((....((((((((....))))))))..((((.---.(((((((..........)))))))..))))---)))---------------)((....((((((....)))))).)).... ( -58.40) >DroEre_CAF1 79163 75 - 1 GACCGUUACACCACCGGAAGCGGUGGUGCCGCCGG---CUCCGGCGACCGUUAUACCACCGGAA------------------------------------------GCGGUGGCGCCAUU (((.(((.((((((((....))))))))..((((.---...))))))).)))...((((((...------------------------------------------.))))))....... ( -38.40) >DroYak_CAF1 67259 75 - 1 GACCGUUACACCACCGGAAACGGUGGUGCUGCCGG---UUCCGGCGAUCGCUAUACCACCGGAA------------------------------------------ACGGUGGCGCCGUU (((((.((((((((((....)))))))).)).)))---)).(((((.........((((((...------------------------------------------.))))))))))).. ( -45.20) >DroAna_CAF1 40584 75 - 1 GGUCGUUAUACCACCGGAAACGGUGGCGGAGUUGG---CUCCGGCGAUCGUUAUACCACCGGAA------------------------------------------ACGGUGGCGGAGUC ((((((....((((((....))))))(((((....---)))))))))))......((((((...------------------------------------------.))))))....... ( -43.60) >consensus GACCGUUACACCACCGGAAACGGUGGUGCCGCCGG___CUCCGGCGAUCGUUAUACCACCGGAA__________________________________________ACGGUGGCGCUGUU ........((((((((....)))))))).((((((.....)))))).........((((((....(((................................)))....))))))....... (-26.36 = -26.52 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:20 2006