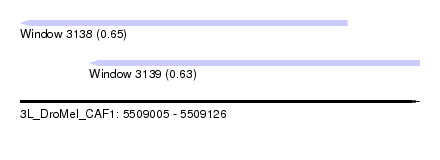
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,509,005 – 5,509,126 |
| Length | 121 |
| Max. P | 0.651825 |

| Location | 5,509,005 – 5,509,104 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -10.07 |
| Energy contribution | -11.35 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5509005 99 - 23771897 UACUAUCCCGGCUCGGGAUAUGGAUCUACCACCUACUACCCAAACCAGGGGAGCUACGGUUCCUCCGGCUACUACCCAACUCAAGGCUAU---------AACUAUUAU ...(((((((...)))))))(((.....))).............((.(((((((....))))))).))......................---------......... ( -24.40) >DroSec_CAF1 71136 99 - 1 UACUAUCCCGGCUCGGGAUAUGGAUCUACCACCUACUAUCCCAACCAGGGGAGCUACGGUUCCUCCGGCUACUACCCAACUCAAGGCUAU---------AACUAUUAU .........((((.((((((.((........))...))))))..((.(((((((....))))))).))................))))..---------......... ( -27.80) >DroSim_CAF1 72614 99 - 1 UACUAUCCCGGGUCGGGAUAUGGAUCUACCACCUACUAUCCCAACCAGGGGAGCUACGGUUCCGCCGGCUACUACCCAACUCAAGGCUAU---------AACUAUUAU .......((.(((.((((((.((........))...)))))).))).))(((((....)))))(((..................)))...---------......... ( -28.87) >DroEre_CAF1 69013 99 - 1 UAUUAUCCAGGCUCGGGAUAUGGCUCUACCACCUACUACCCCAACCAGGGGGGCUAUGGUUCCUCCAGCUACUACCCCACCCAAGGCUAU---------AACUAUUAU .........((...(((...(((((.....(((.....((((......)))).....)))......)))))...)))...))........---------......... ( -21.90) >DroYak_CAF1 60084 93 - 1 UAUUACCCAGGUUCUGGAUACGGAUCUACCGCCUACUAUCCCAACCAGGGCGGCUAUGGUUCC------UACUAUCCAACCCAGGGCUAU---------AACUACUAU (((..(((.(((..((((((.(((.(((((((((.............))))))...))).)))------...)))))))))..)))..))---------)........ ( -27.02) >DroAna_CAF1 31153 97 - 1 -----UCCCAG------CUAUGGCUCUUCCAACUACUAUCCUAACACCGGGUAUUAUGGAUCCUCAAAUGUCUAUUCCAGCCAAGGAUAUGUCCCCAACAAUUACUAC -----......------...(((((...........(((((.......)))))..((((((........))))))...))))).(((....))).............. ( -13.90) >consensus UACUAUCCCGGCUCGGGAUAUGGAUCUACCACCUACUAUCCCAACCAGGGGAGCUACGGUUCCUCCGGCUACUACCCAACCCAAGGCUAU_________AACUAUUAU ...((((((.....))))))(((.....)))(((.....((......))(((((....)))))....................)))...................... (-10.07 = -11.35 + 1.28)

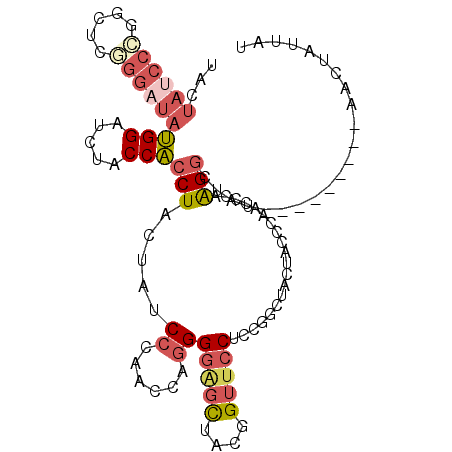
| Location | 5,509,026 – 5,509,126 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.90 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.66 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5509026 100 - 23771897 GAACUACUACGGCUCAUCGGGAUACUAUCCCGGCUCGGGAUAUGGAUCUACCACCUACUACCCAAACCAGGGGAGCUACGGUUCCUCCGGCUACUACCCA ..........((......(((....(((((((...)))))))(((.....))).......)))...((.(((((((....))))))).)).......)). ( -31.40) >DroSec_CAF1 71157 100 - 1 GAACUACUACGGCUCAUCGGGAUACUAUCCCGGCUCGGGAUAUGGAUCUACCACCUACUAUCCCAACCAGGGGAGCUACGGUUCCUCCGGCUACUACCCA ..........((......((((((.(((((((...)))))))(((.....))).....))))))..((.(((((((....))))))).)).......)). ( -34.70) >DroSim_CAF1 72635 100 - 1 GAACUACUACGGCUCAUCGGGAUACUAUCCCGGGUCGGGAUAUGGAUCUACCACCUACUAUCCCAACCAGGGGAGCUACGGUUCCGCCGGCUACUACCCA .........((((....((((((...))))))(((.((((((.((........))...)))))).)))...(((((....)))))))))........... ( -37.90) >DroEre_CAF1 69034 100 - 1 CAACUACUACGGCUCAUCGGGAUAUUAUCCAGGCUCGGGAUAUGGCUCUACCACCUACUACCCCAACCAGGGGGGCUAUGGUUCCUCCAGCUACUACCCC ..........((((.....((((...)))).)))).(((...(((((.....(((.....((((......)))).....)))......)))))...))). ( -24.30) >DroYak_CAF1 60105 94 - 1 GAACUACUACGGCUCAUCGGGCUAUUACCCAGGUUCUGGAUACGGAUCUACCGCCUACUAUCCCAACCAGGGCGGCUAUGGUUCC------UACUAUCCA (((((.(....)......(((......))).))))).(((((.(((.(((((((((.............))))))...))).)))------...))))). ( -27.32) >consensus GAACUACUACGGCUCAUCGGGAUACUAUCCCGGCUCGGGAUAUGGAUCUACCACCUACUAUCCCAACCAGGGGAGCUACGGUUCCUCCGGCUACUACCCA (((((.....(((((...(((....(((((((...)))))))(((.....)))))).....(((.....))))))))..)))))................ (-21.46 = -21.66 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:17 2006