| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 566,819 – 566,910 |
| Length | 91 |
| Max. P | 0.881340 |

| Location | 566,819 – 566,910 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -24.02 |
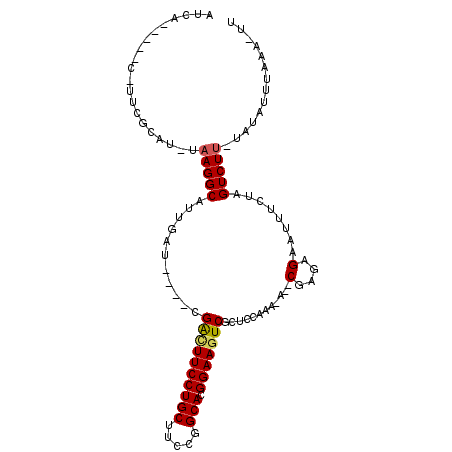
| Consensus MFE | -17.01 |
| Energy contribution | -16.98 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
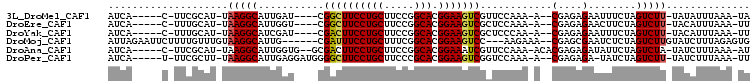
>3L_DroMel_CAF1 566819 91 - 23771897 AUCA-----C-UUCGCAU-UAAGGCAUUGAU----CGGCUUCCUGCUUCCGGCACGGAAGUCGUUCCAAA-A--CGAGAGAAUUUCUAGUCUU-UAUAUUUAAA-UA ....-----.-.......-.(((((...((.----(((((((((((.....))).)))))))).))....-.--(....)........)))))-..........-.. ( -21.60) >DroEre_CAF1 13875 91 - 1 AUCA-----C-UUUGCAU-UAAGGCAUUGGU----CGGCUUCCUGCUUCCGGCACGGAAGUCGCUCCAAA-A--CGAGAGAACUUCUAGUCUU-UACAUUUAAA-UU ....-----.-.......-.(((((.((((.----(((((((((((.....))).))))))))..)))).-.--(....)........)))))-..........-.. ( -25.00) >DroYak_CAF1 14348 91 - 1 AUCA-----C-UUUGCAU-UAAGGCAUCGAU----CGACUUCCUGCUUCCGGCACGGAAGUCGCUCCCAA-A--CGAGAGAAUUUCUAGUCUU-UACAUUUAAA-UU ....-----.-.......-.(((((...((.----(((((((((((.....))).)))))))).))....-.--(....)........)))))-..........-.. ( -22.40) >DroMoj_CAF1 31467 96 - 1 AUUAGAAUUCUUUUGUUUGUAAGGCAUUG------CGAUUUCCUGCUUUCGGCACGGAAGUCC---AAGAAA--CGAGCGAAUCUCUAGUCUUGUAUCUUUAGAGUG ...........(((((((((......(((------.((((((((((.....))).))))))))---))...)--)))))))).((((((..........)))))).. ( -25.00) >DroAna_CAF1 17132 95 - 1 AUCA-----C-UUCGCAU-UAAGGCAUUGGUG--GCGACUUCCUGCUUCCGGCACGGAAAUCGUUCCAAA-ACACGAGAGAUAUUCUAGUCUA-UAUCUUUAAA-AU ....-----.-...((..-....)).((((.(--((((.(((((((.....))).)))).))))))))).-.....((((((((........)-)))))))...-.. ( -24.40) >DroPer_CAF1 64176 94 - 1 AUCA-----U-UUCGCUU-UAAGGCAUUGAGGAUGGGGCUUCCUGCUUCCCGCACGGAAGUCGGUCCAAA-A--CGAGAGA-UAUCUAGUCUU-UAUCUUUAAA-UU ....-----.-...(((.-...))).((((((((((((((((((((.....))).)))))))...))...-.--..(((((-(.....)))))-))))))))).-.. ( -25.70) >consensus AUCA_____C_UUCGCAU_UAAGGCAUUGAU____CGACUUCCUGCUUCCGGCACGGAAGUCGCUCCAAA_A__CGAGAGAAUUUCUAGUCUU_UAUAUUUAAA_UU ....................(((((...........((((((((((.....))).)))))))............(....)........))))).............. (-17.01 = -16.98 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:38 2006