| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,475,624 – 5,475,726 |
| Length | 102 |
| Max. P | 0.850064 |

| Location | 5,475,624 – 5,475,726 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -18.55 |
| Energy contribution | -20.98 |
| Covariance contribution | 2.43 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5475624 102 + 23771897 GUUGACUUGAUUAUCAACACUCGCUGCACAUCUUGCA----UGCUCGC--------------AUCUGUAUCUGAUGCUGCAUGUUGCAGCUGGCAGUUGACAGCUGAUGAUGUUGCAUAU (((((........)))))......((((((((..(((----(((..((--------------(((.......))))).))))))..((((((........))))))..)))).))))... ( -36.50) >DroSec_CAF1 37112 102 + 1 GUUGACUUGAUUAUCAACACUCGCUGCACAUCUUGCA----UGCUCGC--------------AUCUGUAUCUAAUGCUGCAUGUUGCAGCUGGCAGUUGACAGCUGAUGAUGUUGCAUAU (((((........)))))......((((((((..(((----(((..((--------------((.........)))).))))))..((((((........))))))..)))).))))... ( -31.80) >DroSim_CAF1 38798 102 + 1 GUUGACUUGAUUAUCAACACUCGCUGCACAUCUUGCA----UGCUCGC--------------AUCUGUAUCUGAUGCUGCAUGUUGCAGCUGGCAGUUGACAGCUGAUGAUGUUGCAUAU (((((........)))))......((((((((..(((----(((..((--------------(((.......))))).))))))..((((((........))))))..)))).))))... ( -36.50) >DroEre_CAF1 36600 108 + 1 GUUGACUUGAUUAUCAACACUCGCUGCACAUCUUGCA----UGCUCGCAUCU--------GCAUCUGUAUCUGAUGCUGCAUGUUGCAGCUGGCAGUUGACAGCUGAUGAUGUUGCAUAU (((((........)))))......((((((((..(((----(((..((((((--------((....)))...))))).))))))..((((((........))))))..)))).))))... ( -36.40) >DroYak_CAF1 24960 116 + 1 GUUGACUUGAUUAUCAACACUCGCUGCACAUCUUGCA----UGCUCGCAACUCGCAACUCGCAUCUGUAUCUGAUGCAGCAUGUUGCAGCUGGCAGUUGACAGCUGAUGAUGUUGCAUAU (((((........)))))......((((((((..(((----((((.((.....)).....(((((.......))))))))))))..((((((........))))))..)))).))))... ( -38.90) >DroMoj_CAF1 31230 95 + 1 CAUGACUUGAUUAUCACCGCUCGAUGCGC---UUGCACAGCUGAUGGCGACU--------GCAUUUGCAU------UUGCAU-UU-------ACAACUGCCAGCUGAUGAAGUUGCAUGC ((((((((.((((.....((.....))((---(.(((.........((((.(--------((....))).------))))..-..-------.....))).))))))).))))..)))). ( -23.57) >consensus GUUGACUUGAUUAUCAACACUCGCUGCACAUCUUGCA____UGCUCGC______________AUCUGUAUCUGAUGCUGCAUGUUGCAGCUGGCAGUUGACAGCUGAUGAUGUUGCAUAU (((((........)))))......((((((((..(((....)))....................((((..(((..(((((.....)))))...)))...)))).....)))).))))... (-18.55 = -20.98 + 2.43)

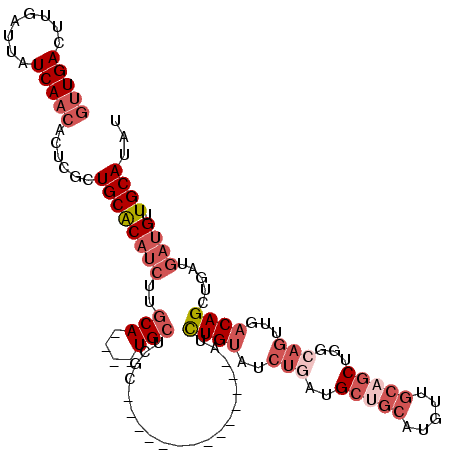
| Location | 5,475,624 – 5,475,726 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -33.71 |
| Consensus MFE | -18.16 |
| Energy contribution | -20.48 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.850064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5475624 102 - 23771897 AUAUGCAACAUCAUCAGCUGUCAACUGCCAGCUGCAACAUGCAGCAUCAGAUACAGAU--------------GCGAGCA----UGCAAGAUGUGCAGCGAGUGUUGAUAAUCAAGUCAAC ...((((.((((..((((((.(....).))))))...(((((.(((((.......)))--------------))..)))----))...))))))))......((((((......)))))) ( -34.90) >DroSec_CAF1 37112 102 - 1 AUAUGCAACAUCAUCAGCUGUCAACUGCCAGCUGCAACAUGCAGCAUUAGAUACAGAU--------------GCGAGCA----UGCAAGAUGUGCAGCGAGUGUUGAUAAUCAAGUCAAC ...((((.((((..((((((.(....).))))))...(((((.(((((.......)))--------------))..)))----))...))))))))......((((((......)))))) ( -32.80) >DroSim_CAF1 38798 102 - 1 AUAUGCAACAUCAUCAGCUGUCAACUGCCAGCUGCAACAUGCAGCAUCAGAUACAGAU--------------GCGAGCA----UGCAAGAUGUGCAGCGAGUGUUGAUAAUCAAGUCAAC ...((((.((((..((((((.(....).))))))...(((((.(((((.......)))--------------))..)))----))...))))))))......((((((......)))))) ( -34.90) >DroEre_CAF1 36600 108 - 1 AUAUGCAACAUCAUCAGCUGUCAACUGCCAGCUGCAACAUGCAGCAUCAGAUACAGAUGC--------AGAUGCGAGCA----UGCAAGAUGUGCAGCGAGUGUUGAUAAUCAAGUCAAC ...((((.((((..((((((.(....).))))))...(((((.(((((............--------.)))))..)))----))...))))))))......((((((......)))))) ( -34.02) >DroYak_CAF1 24960 116 - 1 AUAUGCAACAUCAUCAGCUGUCAACUGCCAGCUGCAACAUGCUGCAUCAGAUACAGAUGCGAGUUGCGAGUUGCGAGCA----UGCAAGAUGUGCAGCGAGUGUUGAUAAUCAAGUCAAC ...((((((.((...(((((.(....).)))))(((((....((((((.......)))))).))))))))))))).((.----((((.....))))))....((((((......)))))) ( -37.00) >DroMoj_CAF1 31230 95 - 1 GCAUGCAACUUCAUCAGCUGGCAGUUGU-------AA-AUGCAA------AUGCAAAUGC--------AGUCGCCAUCAGCUGUGCAA---GCGCAUCGAGCGGUGAUAAUCAAGUCAUG ((.((((.......((((((((..((((-------(.-.(((..------..)))..)))--------))..)))...))))))))).---))((.....)).(((((......))))). ( -28.61) >consensus AUAUGCAACAUCAUCAGCUGUCAACUGCCAGCUGCAACAUGCAGCAUCAGAUACAGAU______________GCGAGCA____UGCAAGAUGUGCAGCGAGUGUUGAUAAUCAAGUCAAC ...((((.((((.....((((...(((...(((((.....)))))..)))..))))....................(((....)))..))))))))......((((((......)))))) (-18.16 = -20.48 + 2.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:07 2006