
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,460,998 – 5,461,093 |
| Length | 95 |
| Max. P | 0.999661 |

| Location | 5,460,998 – 5,461,093 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -18.32 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
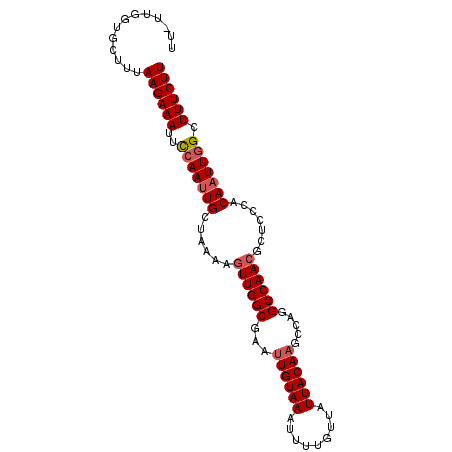
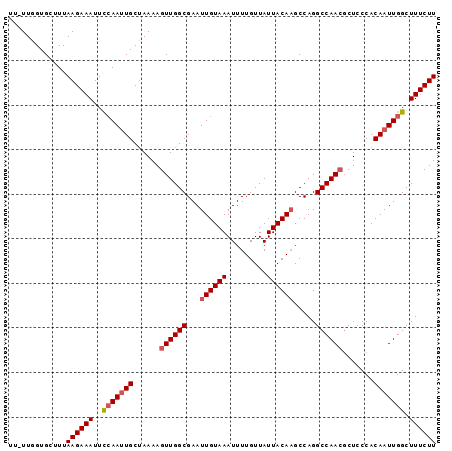
>3L_DroMel_CAF1 5460998 95 + 23771897 UU-UUGGUGCUUUAAGAAAUUCCAAUUGCUAAAAGUUGGCGAAUUGUAAAUUUUGUUAUUACAAGCCAGGCCAACUCUCCCACAAUUGGCUUUCUU ..-..........((((((..(((((((.....(((((((...((((((.........)))))).....)))))))......))))))).)))))) ( -22.90) >DroSec_CAF1 22442 95 + 1 UU-UUUGUGCUUUAAGAAAUUUCAAUUGCGAAAAGUUGGCGAAUUGUAAAUUUUGUUAUUACACGCCAGGCCAAAGCUCCCACAAUUGGCUUUCUU ..-..........((((((..(((((((.(......(((((...(((((.........))))))))))(((....)))..).))))))).)))))) ( -18.20) >DroSim_CAF1 23887 96 + 1 UUUUUUGUGCUUUAAGAAAUUUCAAUUGCUAAAAGUUGGCGAAUUGUAAAUUUUGUUAUUACAAGCCAGGCCAACGCUCCCACAAUUGGCUUUCUU .............((((((..(((((((......((((((...((((((.........)))))).....)))))).......))))))).)))))) ( -19.92) >DroEre_CAF1 22089 94 + 1 UC-UUAGUGCUUUAAGAAAUUCCAA-UGCUAAAAGUUGGCGAAUUGUAAAUUUUGUUAUUACAAGCCAGGCCAACGCUCCCACAAUUGGCUUUCUU ..-..........((((((..((((-(.......((((((...((((((.........)))))).....)))))).........))))).)))))) ( -19.09) >DroYak_CAF1 9258 95 + 1 UU-UUGGUGCUUUAAGAAAUUCAAAUUGCUAAAAGUUGGCGAAUUGUAAAUUUUGUUAUUACAAGCCAGGCCAACGCUCCCACAAUUGGCUUUCUU ..-..........((((((..(((.(((......((((((...((((((.........)))))).....)))))).......))))))..)))))) ( -18.22) >consensus UU_UUGGUGCUUUAAGAAAUUCCAAUUGCUAAAAGUUGGCGAAUUGUAAAUUUUGUUAUUACAAGCCAGGCCAACGCUCCCACAAUUGGCUUUCUU .............((((((..(((((((......((((((...((((((.........)))))).....)))))).......))))))).)))))) (-18.32 = -18.88 + 0.56)

| Location | 5,460,998 – 5,461,093 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5460998 95 - 23771897 AAGAAAGCCAAUUGUGGGAGAGUUGGCCUGGCUUGUAAUAACAAAAUUUACAAUUCGCCAACUUUUAGCAAUUGGAAUUUCUUAAAGCACCAA-AA ((((((.((((((((..((((((((((..(..((((((.........))))))..))))))))))).))))))))..))))))..........-.. ( -32.30) >DroSec_CAF1 22442 95 - 1 AAGAAAGCCAAUUGUGGGAGCUUUGGCCUGGCGUGUAAUAACAAAAUUUACAAUUCGCCAACUUUUCGCAAUUGAAAUUUCUUAAAGCACAAA-AA ((((((..((((((((..(((....)).((((((((((.........)))))...)))))...)..))))))))...))))))..........-.. ( -25.60) >DroSim_CAF1 23887 96 - 1 AAGAAAGCCAAUUGUGGGAGCGUUGGCCUGGCUUGUAAUAACAAAAUUUACAAUUCGCCAACUUUUAGCAAUUGAAAUUUCUUAAAGCACAAAAAA ((((((..(((((((..(((.((((((..(..((((((.........))))))..))))))))))..)))))))...))))))............. ( -25.70) >DroEre_CAF1 22089 94 - 1 AAGAAAGCCAAUUGUGGGAGCGUUGGCCUGGCUUGUAAUAACAAAAUUUACAAUUCGCCAACUUUUAGCA-UUGGAAUUUCUUAAAGCACUAA-GA ((((((.(((((.((..(((.((((((..(..((((((.........))))))..))))))))))..)))-))))..))))))..........-.. ( -23.70) >DroYak_CAF1 9258 95 - 1 AAGAAAGCCAAUUGUGGGAGCGUUGGCCUGGCUUGUAAUAACAAAAUUUACAAUUCGCCAACUUUUAGCAAUUUGAAUUUCUUAAAGCACCAA-AA ((((((..(((((((..(((.((((((..(..((((((.........))))))..))))))))))..)))).)))..))))))..........-.. ( -23.20) >consensus AAGAAAGCCAAUUGUGGGAGCGUUGGCCUGGCUUGUAAUAACAAAAUUUACAAUUCGCCAACUUUUAGCAAUUGGAAUUUCUUAAAGCACAAA_AA ((((((..(((((((..(((.((((((..((.((((((.........)))))).)))))))))))..)))))))...))))))............. (-22.50 = -23.30 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:50 2006