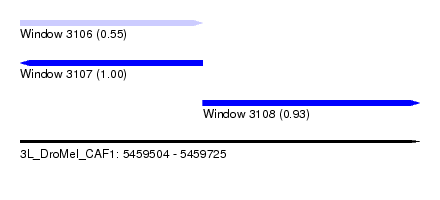
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,459,504 – 5,459,725 |
| Length | 221 |
| Max. P | 0.999439 |

| Location | 5,459,504 – 5,459,605 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.52 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -15.75 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5459504 101 + 23771897 CAAAGUUCAGGCCUUUCAAUGUUUUUUUUUGCCUUGUUUUUUUGCUGUAUUAAAUGGCAAAAUUUCAUUGCGAAAAUUGGUAACUUUGAAAUAUUUACAUU (((((((...(((............(((((((..((...(((((((((.....)))))))))...))..)))))))..))))))))))............. ( -21.54) >DroSec_CAF1 21010 97 + 1 CAAAGUUCAGGCUUUUCAAUGUUU--UUUUGCCUUGUU--UGUGCUGUAUUAAAUGGCAAAAUUUAAUUGCGAAAAUUGGUAACUUUGAAAUAUUUAGAUU ((((((((((..(((.((((....--(((((((..(((--((........))))))))))))....)))).)))..))))..))))))............. ( -19.50) >DroSim_CAF1 22417 97 + 1 CAAAGUUCAGGCCUUUUAAUGAUU--UUUUGCCUUGUU--UGUGCUGUAUUAAAUGGCAAAAUUUCAUUGCGAAAAUUGGUAACUUUGAAAUAUUUACAUU ((((((((((...((.((((((..--(((((((..(((--((........))))))))))))..)))))).))...))))..))))))............. ( -21.80) >DroEre_CAF1 20527 96 + 1 CAAAGUUCAGGCCUUUCGAUGUUU---UUUGCCUUGUU--UGUGCUGUAUUAAAUGGCAAAAUUUCAUUGCGAAAAUUGGUAACUUUGAAAUAUUUACAUU (((((((...(((((((((((..(---((((((..(((--((........))))))))))))...)))..)))))...))))))))))............. ( -20.90) >DroYak_CAF1 7696 97 + 1 CAAAGUUCAGGCCUUUCGAUGUUU--UUUUGCCAUGUU--UGUGCUGUAUUAAAUGGCAAAAUUUCAUUGCGAAAAUUGGUAACUUUGAAAUAUUUACAUU (((((((...(((((((((((...--(((((((((...--.............)))))))))...)))..)))))...))))))))))............. ( -22.39) >consensus CAAAGUUCAGGCCUUUCAAUGUUU__UUUUGCCUUGUU__UGUGCUGUAUUAAAUGGCAAAAUUUCAUUGCGAAAAUUGGUAACUUUGAAAUAUUUACAUU ((((((((((...((.(((((.....(((((((......................)))))))...))))).))...))))..))))))............. (-15.75 = -15.75 + 0.00)

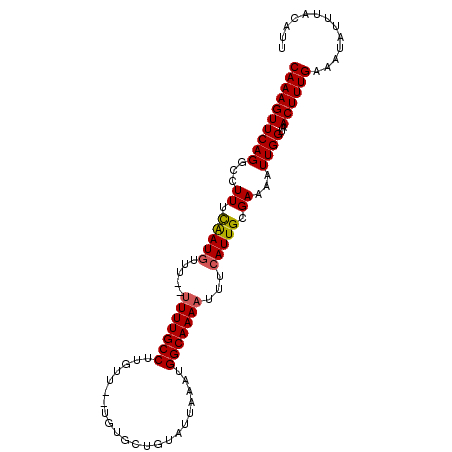
| Location | 5,459,504 – 5,459,605 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.52 |
| Mean single sequence MFE | -16.34 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5459504 101 - 23771897 AAUGUAAAUAUUUCAAAGUUACCAAUUUUCGCAAUGAAAUUUUGCCAUUUAAUACAGCAAAAAAACAAGGCAAAAAAAAACAUUGAAAGGCCUGAACUUUG .............(((((((.....((((((........(((((((......................)))))))........)))))).....))))))) ( -14.94) >DroSec_CAF1 21010 97 - 1 AAUCUAAAUAUUUCAAAGUUACCAAUUUUCGCAAUUAAAUUUUGCCAUUUAAUACAGCACA--AACAAGGCAAAA--AAACAUUGAAAAGCCUGAACUUUG .............(((((((.....((((((........(((((((...............--.....)))))))--......)))))).....))))))) ( -14.49) >DroSim_CAF1 22417 97 - 1 AAUGUAAAUAUUUCAAAGUUACCAAUUUUCGCAAUGAAAUUUUGCCAUUUAAUACAGCACA--AACAAGGCAAAA--AAUCAUUAAAAGGCCUGAACUUUG .............(((((((.....((((...(((((..(((((((...............--.....)))))))--..))))).)))).....))))))) ( -15.25) >DroEre_CAF1 20527 96 - 1 AAUGUAAAUAUUUCAAAGUUACCAAUUUUCGCAAUGAAAUUUUGCCAUUUAAUACAGCACA--AACAAGGCAAA---AAACAUCGAAAGGCCUGAACUUUG .............(((((((.....((((((..(((...(((((((...............--.....))))))---)..))))))))).....))))))) ( -17.35) >DroYak_CAF1 7696 97 - 1 AAUGUAAAUAUUUCAAAGUUACCAAUUUUCGCAAUGAAAUUUUGCCAUUUAAUACAGCACA--AACAUGGCAAAA--AAACAUCGAAAGGCCUGAACUUUG .............(((((((.....((((((..(((...(((((((((.............--...)))))))))--...))))))))).....))))))) ( -19.69) >consensus AAUGUAAAUAUUUCAAAGUUACCAAUUUUCGCAAUGAAAUUUUGCCAUUUAAUACAGCACA__AACAAGGCAAAA__AAACAUUGAAAGGCCUGAACUUUG .............(((((((.....((((((........(((((((......................)))))))........)))))).....))))))) (-15.18 = -15.18 + -0.00)

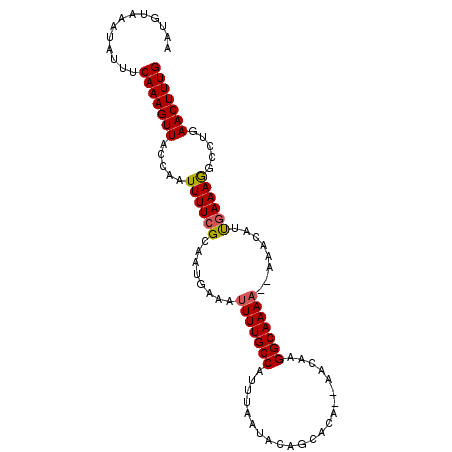
| Location | 5,459,605 – 5,459,725 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -20.98 |
| Energy contribution | -20.46 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5459605 120 + 23771897 UUAGUAACAUUUCAUAAAAAGCUUAUGCAAAAUAAAUGGGUAUGGAAAAGCCGGAAAAGUGGUUUACUGGUAUUUAUGGGAACAUAAUCAAAUGUUAUAGCCAGUUAGGGGCAAAUGCAA ....................((...(((....(((.(((.(((((....(((((.(((....))).)))))..(((((....))))).......))))).))).)))...)))...)).. ( -23.40) >DroSec_CAF1 21107 120 + 1 UCGGUAAAAUUUCAUAAAAAGCUUAUGAAAAAUAAAUGGGUAUGGAAAUGCCGGAAUAGUGGUUUACUGGUAUUUAUGAAAACAUAAUCAAAUGUUAUGGCCAGUUAGGGGCAAGUGCAA .(((((((.((((((((.....))))))))........(((((....)))))..........)))))))((((((.......((((((.....))))))(((.......))))))))).. ( -29.00) >DroSim_CAF1 22514 120 + 1 UCGGUAAAAUUUCAUAAAAAGCUUAUGCAAAAUAAAUGGGUAUGGAAAAGCCGGAAAAGUGGUUUACUGGUAUUUAUGAAAACAUAAUCAAAUGUUAUGGCCAGUUAGGGGUAAAUGCAA .(((((((.((((.......((....))..........(((.(....).))).)))).....)))))))((((((((.....((((((.....)))))).((......)))))))))).. ( -22.60) >DroEre_CAF1 20623 120 + 1 UCAGUAAAAUUUCAUAAAAAGCUUAUGCAAAAUAAAUGGGUAUGGAAAAGCCGGAAAAGUGGUUUACUGGUAUUUAUGAAAACAUAAUCAAAUGUUAUGGCCAGUUUGGGGCAAAUGCAA .(((((((.((((.......((....))..........(((.(....).))).)))).....)))))))((((((.......((((((.....))))))(((.......))))))))).. ( -23.60) >DroYak_CAF1 7793 120 + 1 UCACUAAAAUUUCAUAAAAAGCUUAUGCAAAAUAAAUGGGUAUGGAAAAGCCGGAAAAGUGGUUUACUGGUAUUUAUGAAAACAUAAUCAAAUGUUAUAGCCAGUUUGGGGCAAAUGCAA .........((((((.....((((((.........))))))))))))..(((...(((.(((((.((......(((((....)))))......))...))))).)))..)))........ ( -20.50) >consensus UCAGUAAAAUUUCAUAAAAAGCUUAUGCAAAAUAAAUGGGUAUGGAAAAGCCGGAAAAGUGGUUUACUGGUAUUUAUGAAAACAUAAUCAAAUGUUAUGGCCAGUUAGGGGCAAAUGCAA .((((((..((((((.....((((((.........))))))))))))..((((......))))))))))((((((.....(((((......)))))...(((.......))))))))).. (-20.98 = -20.46 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:47 2006