
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,449,324 – 5,449,456 |
| Length | 132 |
| Max. P | 0.987791 |

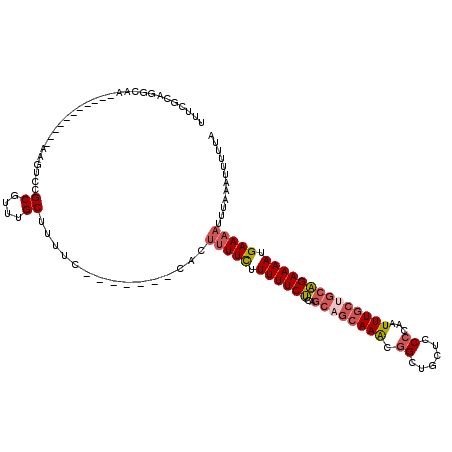
| Location | 5,449,324 – 5,449,426 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -12.72 |
| Energy contribution | -13.88 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5449324 102 + 23771897 AUUCGCAGGCAA-----------AAGUCCGCGUUUGCUUUUC-------CACUUUUCUUUUUCUCAGCAGCAAACGGCUGCUCCCCAAUUUGCUGCAGAAAAUGAAAAUUUAAAUUUUUA ...(((.(((..-----------..))).)))..........-------...(((((.((((((..((((((((.((......))...)))))))))))))).)))))............ ( -24.20) >DroSec_CAF1 10758 102 + 1 UUUCGCAGGCAA-----------AAGUCCGCGUUUGCUUUUC-------CACUUUUCUUUUUCUCAGCAGCAAACGGCUGCUCCCCAAUUUGCUGCAGAAAAUGAAAAUUUAAAUUUUUA ...(((.(((..-----------..))).)))..........-------...(((((.((((((..((((((((.((......))...)))))))))))))).)))))............ ( -24.20) >DroSim_CAF1 10858 102 + 1 UCUCGCAGGCAA-----------AAGUCCGCGUUUGCUUUUC-------CACUUUUCUUUUUCUCAGCAGCAAACGGCUGCUCCCCAAUUUGCUGCAGAAAAUGAAAAUUUAAAUUUUUA ...(((.(((..-----------..))).)))..........-------...(((((.((((((..((((((((.((......))...)))))))))))))).)))))............ ( -24.20) >DroEre_CAF1 10507 98 + 1 -----------A-----------AAGUCCGCGUUUGCUUUUCCACAUUCCACUUUUCUUUUUCUCAGCAGCAAACUGCUGCUCCCCAAUUUGCAGCAGAAAAUGAAAAUUUAAAUUUUUA -----------.-----------........(((((((..............................)))))))((((((..........))))))..((((....))))......... ( -15.21) >DroMoj_CAF1 1644 96 + 1 CUUUGCGAGCAACUUUAGCCCAGGAGGCGGCAGCUGCU---------------GUUUUUUUUCC-------AAGAGGUU--UUCCCAAUUUGCGGCGGAAAAUGAAAAUUUAAAUUUUUA .(((((..((((.........((((((((((....)))---------------)))))))....-------....((..--...))...)))).)))))((((....))))......... ( -18.50) >consensus UUUCGCAGGCAA___________AAGUCCGCGUUUGCUUUUC_______CACUUUUCUUUUUCUCAGCAGCAAACGGCUGCUCCCCAAUUUGCUGCAGAAAAUGAAAAUUUAAAUUUUUA .............................((....))...............(((((.((((((..((((((((.((......))...)))))))))))))).)))))............ (-12.72 = -13.88 + 1.16)


| Location | 5,449,324 – 5,449,426 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -14.32 |
| Energy contribution | -16.40 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5449324 102 - 23771897 UAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG-------GAAAAGCAAACGCGGACUU-----------UUGCCUGCGAAU ......(((...(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))...-------))).......(((((.(..-----------..).)))))... ( -29.10) >DroSec_CAF1 10758 102 - 1 UAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG-------GAAAAGCAAACGCGGACUU-----------UUGCCUGCGAAA ......(((...(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))...-------))).......(((((.(..-----------..).)))))... ( -29.10) >DroSim_CAF1 10858 102 - 1 UAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG-------GAAAAGCAAACGCGGACUU-----------UUGCCUGCGAGA ......(((...(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))...-------))).......(((((.(..-----------..).)))))... ( -29.10) >DroEre_CAF1 10507 98 - 1 UAAAAAUUUAAAUUUUCAUUUUCUGCUGCAAAUUGGGGAGCAGCAGUUUGCUGCUGAGAAAAAGAAAAGUGGAAUGUGGAAAAGCAAACGCGGACUU-----------U----------- ............(((((((.(((..((.....((......((((((....)))))).....))....))..))).))))))).((....))......-----------.----------- ( -23.60) >DroMoj_CAF1 1644 96 - 1 UAAAAAUUUAAAUUUUCAUUUUCCGCCGCAAAUUGGGAA--AACCUCUU-------GGAAAAAAAAC---------------AGCAGCUGCCGCCUCCUGGGCUAAAGUUGCUCGCAAAG ..................(((((((........))))))--).((....-------)).........---------------(((((((...(((.....)))...)))))))....... ( -19.80) >consensus UAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG_______GAAAAGCAAACGCGGACUU___________UUGCCUGCGAAA ............(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))...............((....))............................. (-14.32 = -16.40 + 2.08)



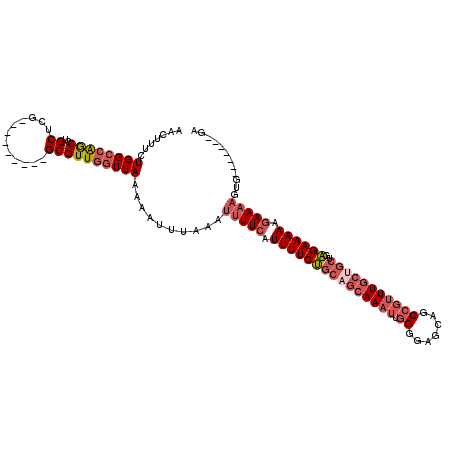
| Location | 5,449,353 – 5,449,456 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -19.20 |
| Energy contribution | -21.36 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5449353 103 - 23771897 AACUUUCUGGCCAGCCU-GCUCG---------GCGUUGGUUAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG-------GA .......((((((((((-....)---------).))))))))..........(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))...-------.. ( -31.30) >DroSec_CAF1 10787 103 - 1 AACUUUCUGGCCAGCCU-GCUCG---------GCGUUGGUUAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG-------GA .......((((((((((-....)---------).))))))))..........(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))...-------.. ( -31.30) >DroSim_CAF1 10887 103 - 1 AACUUUCUGGCCAGCCU-GCUCG---------GCGUUGGUUAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG-------GA .......((((((((((-....)---------).))))))))..........(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))...-------.. ( -31.30) >DroEre_CAF1 10525 110 - 1 AACUUUCUGGCCAGCCG-GCUCG---------GCGUUGGUUAAAAAUUUAAAUUUUCAUUUUCUGCUGCAAAUUGGGGAGCAGCAGUUUGCUGCUGAGAAAAAGAAAAGUGGAAUGUGGA .......((((((((((-...))---------))..))))))...((((...(((((.((((((((.((((((((........)))))))).))..)))))).)))))...))))..... ( -30.60) >DroMoj_CAF1 1682 98 - 1 AACUUUUUGGCAGACUUCGCUCGUCGACGUUUGCGUUGCUUAAAAAUUUAAAUUUUCAUUUUCCGCCGCAAAUUGGGAA--AACCUCUU-------GGAAAAAAAAC------------- .........((((((.(((.....))).))))))........................(((((((........))))))--).((....-------)).........------------- ( -15.70) >consensus AACUUUCUGGCCAGCCU_GCUCG_________GCGUUGGUUAAAAAUUUAAAUUUUCAUUUUCUGCAGCAAAUUGGGGAGCAGCCGUUUGCUGCUGAGAAAAAGAAAAGUG_______GA .......((((((((...((............))))))))))..........(((((.(((((((((((((((.((.......)))))))))))..)))))).)))))............ (-19.20 = -21.36 + 2.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:30 2006