
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,447,296 – 5,447,455 |
| Length | 159 |
| Max. P | 0.789883 |

| Location | 5,447,296 – 5,447,416 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -23.44 |
| Energy contribution | -22.88 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5447296 120 - 23771897 GUGAAUAUUAAUGCCUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCUUACUUCAUGACCCUGGACGCGAUUCGUGAUGCUCUCAGACCACUUAGGCAAAUGUUUAUGAUUUU (((((((((..(((((((..........((((.....((((.((((...(((....)))....)))).))))((((...)))).))))..........))))))))))))))))...... ( -27.15) >DroSec_CAF1 8836 120 - 1 GUGAAUAUUAAUGCUUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCCUACUUCAUGACCCUGGACGCGAUUCGGUAUGCCCUCAGACCACUUAAGCAAAUGUUUAUGAUUUU (((((((((..(((((((..........(((((((....((.((((...(((....)))....)))).))((......))))).))))..........))))))))))))))))...... ( -25.85) >DroSim_CAF1 8887 120 - 1 GUGAAUAUUAAUGCCUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCUUACUGCAUGACCCUGGACGCGAUUCGGUAUGCCCUCAGACCACUUAGGCAAAUGUUUAUGAUUUU (((((((((..(((((((..........(((((((....((.((((..((((....))))...)))).))((......))))).))))..........))))))))))))))))...... ( -28.95) >DroEre_CAF1 8636 120 - 1 GUGAAUAUUAAUGCCAAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUUUUGCUCCAUGACCCUGGACGCGAUUCGCUAUGCUCUCAGACCACUUAGGCAAAUGUUUAUGAUUUU (((((((((..((((.............((((((((.....))))...((((..((((((((......)))).))))...)))))))).............)))))))))))))...... ( -25.87) >consensus GUGAAUAUUAAUGCCUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCUUACUUCAUGACCCUGGACGCGAUUCGGUAUGCCCUCAGACCACUUAGGCAAAUGUUUAUGAUUUU (((((((((..(((((((..........((((.....((((.((((...(((....)))....)))).))))((.....))...))))..........))))))))))))))))...... (-23.44 = -22.88 + -0.56)

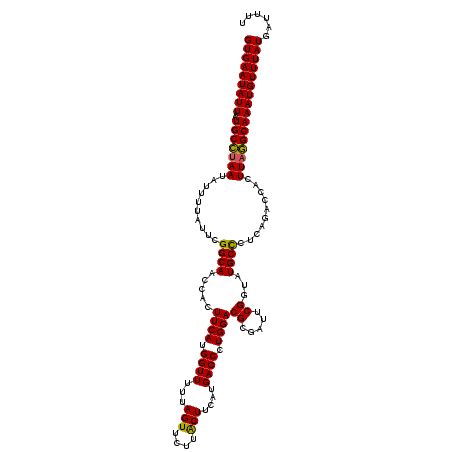

| Location | 5,447,336 – 5,447,455 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -25.77 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5447336 119 + 23771897 GAAUCGCGUCCAGGGUCAUGAAGUAAGAACUAAAAACCAUGAAGUGGUUGCCGAAUAAAAUAUUAGGCAUUAAUAUUCACACCCUUAGAGGGCUCACGACGACGGGGUCAUGACC-CAUC ............(((((((((.....(((.....((((((...))))))(((.((((...)))).))).......)))...(((.....)))(((.((....)))))))))))))-)... ( -31.20) >DroSec_CAF1 8876 117 + 1 GAAUCGCGUCCAGGGUCAUGAAGUAGGAACUAAAAACCAUGAAGUGGUUGCCGAAUAAAAUAUUAAGCAUUAAUAUUCACACCCUUCGAGGACUCACGACG---GGGUCAUGACCCCACC ............(((((((((.((.(((......((((((...))))))((..((((...))))..)).......))))).(((.(((........))).)---)).))))))))).... ( -30.10) >DroSim_CAF1 8927 120 + 1 GAAUCGCGUCCAGGGUCAUGCAGUAAGAACUAAAAACCAUGAAGUGGUUGCCGAAUAAAAUAUUAGGCAUUAAUAUUCACACCCUUCGAGGACUCACGACGGUGGGGUCAUGACCCCACC ...(((.(((((((((..((.((((.........((((((...))))))(((.((((...)))).))).....)))))).)))))....))))...))).((((((((....)))))))) ( -37.60) >DroEre_CAF1 8676 116 + 1 GAAUCGCGUCCAGGGUCAUGGAGCAAAAACUAAAAACCAUGAAGUGGUUGCCGAAUAAAAUAUUUGGCAUUAAUAUUCACACCCUUCGAGGGCUCACGACG---GGGUCAUGACCCGAC- .......(((..(((((((((((...........((((((...))))))((((((((...)))))))).......)))..((((((((........))).)---)))))))))))))))- ( -36.10) >consensus GAAUCGCGUCCAGGGUCAUGAAGUAAGAACUAAAAACCAUGAAGUGGUUGCCGAAUAAAAUAUUAGGCAUUAAUAUUCACACCCUUCGAGGACUCACGACG___GGGUCAUGACCCCACC ............((((((((................((.(((((.(((((((.((((...)))).))))...........)))))))).))((((.........)))))))))))).... (-25.77 = -26.28 + 0.50)

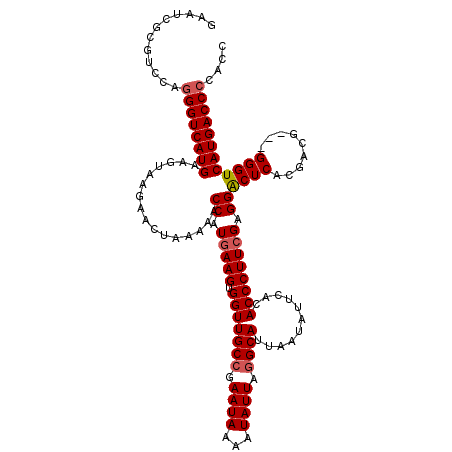
| Location | 5,447,336 – 5,447,455 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -29.24 |
| Energy contribution | -30.43 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5447336 119 - 23771897 GAUG-GGUCAUGACCCCGUCGUCGUGAGCCCUCUAAGGGUGUGAAUAUUAAUGCCUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCUUACUUCAUGACCCUGGACGCGAUUC ((((-((.......))))))(((((((((((.....))))((((..(((((((((.((((...)))).))))((((.....))))..)))))..)))).))))))).............. ( -34.40) >DroSec_CAF1 8876 117 - 1 GGUGGGGUCAUGACCC---CGUCGUGAGUCCUCGAAGGGUGUGAAUAUUAAUGCUUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCCUACUUCAUGACCCUGGACGCGAUUC ...(((((....))))---)(((((..((((..((((((((((((((..((((.....)))).))))).)).))).))))..((((...(((....)))....))))..))))))))).. ( -34.90) >DroSim_CAF1 8927 120 - 1 GGUGGGGUCAUGACCCCACCGUCGUGAGUCCUCGAAGGGUGUGAAUAUUAAUGCCUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCUUACUGCAUGACCCUGGACGCGAUUC ((((((((....))))))))(((((..((((..((((((((((((((..((((.....)))).))))).)).))).))))..((((..((((....))))...))))..))))))))).. ( -43.20) >DroEre_CAF1 8676 116 - 1 -GUCGGGUCAUGACCC---CGUCGUGAGCCCUCGAAGGGUGUGAAUAUUAAUGCCAAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUUUUGCUCCAUGACCCUGGACGCGAUUC -(((((((((((((((---..(((.(....).))).))))(..((.(((((((((.((((...)))).))))((((.....))))..))))).))..)..))))))))..)))....... ( -36.60) >consensus GGUGGGGUCAUGACCC___CGUCGUGAGCCCUCGAAGGGUGUGAAUAUUAAUGCCUAAUAUUUUAUUCGGCAACCACUUCAUGGUUUUUAGUUCUUACUUCAUGACCCUGGACGCGAUUC ((((((((....))))))))((((((.((((.....))))...........((((.((((...)))).)))).....((((.((((...(((....)))....)))).)))))))))).. (-29.24 = -30.43 + 1.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:26 2006