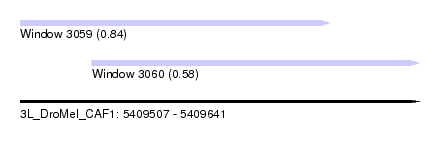
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,409,507 – 5,409,641 |
| Length | 134 |
| Max. P | 0.839086 |

| Location | 5,409,507 – 5,409,611 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
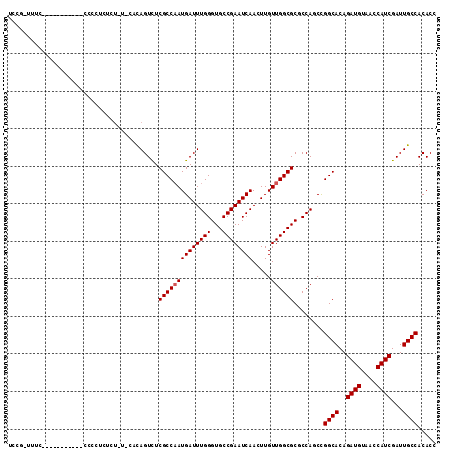
>3L_DroMel_CAF1 5409507 104 + 23771897 UCCG-UUUCCA---------UCUCUCUUUAUCCACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACG ....-......---------..............(((((..(((...(((..(((((((((.........))))))))).))).)))...((((....)))))))))....... ( -27.50) >DroVir_CAF1 28018 98 + 1 UCUG----------------CCCCGCGCUUUAUUCAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACC ...(----------------(..((((((......)))...(((((((((((((...)))))))).....))))))))...))(((((..((((....))))...))))).... ( -28.30) >DroGri_CAF1 55957 103 + 1 ACUUCGCUU-----------CGCGUCUCUUCAAAUAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACC .....(((.-----------((((...((......))...)(((((((((((((...)))))))).....))))))))..)))(((((..((((....))))...))))).... ( -29.70) >DroYak_CAF1 4617 113 + 1 UCCG-UUUCCCUCUCUUAUUCCCCUCUCUAUCCACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCUCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACG ....-.............................(((((..(((...(((..(((((((((.........))))))))).))).)))...((((....)))))))))....... ( -24.50) >DroMoj_CAF1 27474 99 + 1 UCUC-CAUU-----------CACCUCUCU---UGCAGUCUCGCCCAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACC ....-....-----------.........---.((((((..(((..((((((((...))))))))....(((((....))))).)))...((((....))))))))))...... ( -29.30) >DroAna_CAF1 20069 91 + 1 UUCA-UUUCAA----------------------ACAGUUUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACG ....-......----------------------..............(((..(((((((((.........))))))))).))).((((..((((....))))...))))..... ( -25.00) >consensus UCCG_UUUC___________CCCCUCUCU_U_CACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCCGGCACAGAUGUAACCAUCGAUUGCCACACC ........................................((((((((((((((...)))))))).....))))))........((((..((((....))))...))))..... (-23.48 = -23.65 + 0.17)


| Location | 5,409,531 – 5,409,641 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -29.35 |
| Energy contribution | -28.93 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5409531 110 + 23771897 CAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCGCAUCCCAAGUGAGUCUACUC-GGA-U-------- .((.(((.....((((((((...))))))))(((((...(((((((....((((..((((....))))...))))....)).)))))....)))))))).))....-...-.-------- ( -39.40) >DroVir_CAF1 28036 112 + 1 CAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCUCAUCCCAAGUGAGUUUUUCAUUCAUU-------- .(((((((....((((((((...))))))))....(((((....)))))(((((..((((....))))...)))))...))))))).......(((((((.....)))))))-------- ( -35.40) >DroGri_CAF1 55980 109 + 1 UAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCGCAUCCCAAGUAAGCUUUUCAUA---U-------- .......((.....(((((((.(((((.........)))))(((((.(.(((((..((((....))))...)))))...)..)))))..)))))))..))........---.-------- ( -34.40) >DroWil_CAF1 73871 118 + 1 CAGUCUCUCCCAUGAUUUGGGUACCGAAUCAACUUGUUGGUGCUCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCACAUCCCAAGUAAGUUCUUUA-CAC-UUUUCUCUC ..(((((.....((((((((...))))))))....(((((....)))))(((((..((((....))))...)))))....))))).........(((((...))))-)..-......... ( -30.70) >DroMoj_CAF1 27493 112 + 1 CAGUCUCGCCCAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCCCAUCCCAAGUGAGUUUUUCUCCCACU-------- ..((((((....((((((((...))))))))....(((((....)))))(((((..((((....))))...)))))...)))))).........(.(((.....))).)...-------- ( -33.40) >DroAna_CAF1 20080 117 + 1 CAGUUUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCACAUCCCAAGUGAGUUUUCCCUAGA-U-UUUCAU- .......(((........(((((((((.........)))))))))..(((((((..((((....))))...))))....)))))).........(((((...((....))-.-.)))))- ( -33.00) >consensus CAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCACAUCCCAAGUGAGUUUUUCAUACA_U________ ..(((((..........((((((((((.........))))))).)))...((((..((((....))))...)))).....)))))................................... (-29.35 = -28.93 + -0.42)

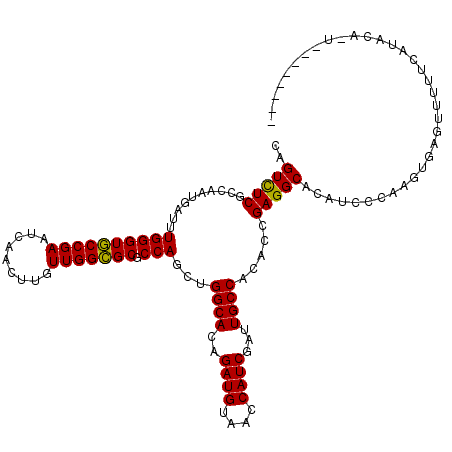

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:01 2006