| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,404,718 – 5,404,831 |
| Length | 113 |
| Max. P | 0.998689 |

| Location | 5,404,718 – 5,404,831 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5404718 113 + 23771897 AAACGGAGACAAAAGUGGCAUCAGGCAGCCUCAACAGGCAGCAGCUACGUGCUGCCACU-UUGCCCCACC------CACCAUGCCACUUCCACAUUGCACUCCCCGCUCUCCCCCCCCCC ....(((((...(((((((((..((((((((....)))).(((((.....)))))....-.)))).....------....))))))))).......((.......)))))))........ ( -36.10) >DroSim_CAF1 50550 112 + 1 AAACGGAGACAAAAGUGGCAUCAGGCAGCCUCAACAGGCAGCAGCUACGUGCUGCCACU-UUGCCCCACCGCCCGCCAACAUGCCACUUUCCCAUUGCACUCCCCACUG-------CCCC ....((((.((((((((((((..(((.((((....)))).(((((.....)))))....-..............)))...))))))))).....)))..))))......-------.... ( -36.70) >DroYak_CAF1 53748 113 + 1 AAACGGAGACAAAAGUGGCAUCAGGCAGCCUCAACAGGCAGCAGCUACGUGCUGCCACUGCUGCCCCACCGCU--CCAUCAUGCCACUUCCAUGCCCCAAC-----ACCACCCCCACCCC ....((.(.((.(((((((((..((((((.......(((((((......)))))))...)))))).....(..--....))))))))))...))).))...-----.............. ( -34.20) >consensus AAACGGAGACAAAAGUGGCAUCAGGCAGCCUCAACAGGCAGCAGCUACGUGCUGCCACU_UUGCCCCACCGC___CCAACAUGCCACUUCCACAUUGCACUCCCC_CUC_CCCCC_CCCC ....(....)..(((((((((..((((((((....)))).(((((.....)))))......))))...............)))))))))............................... (-30.40 = -30.40 + -0.00)

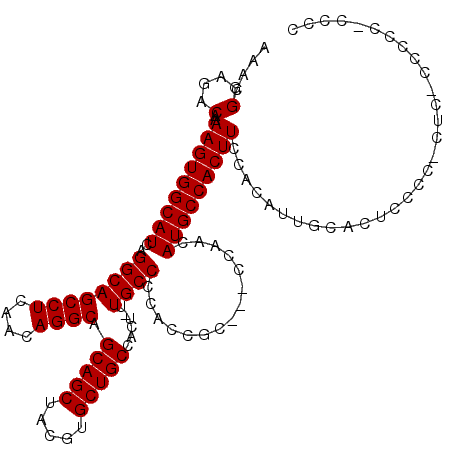
| Location | 5,404,718 – 5,404,831 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -48.17 |
| Consensus MFE | -37.57 |
| Energy contribution | -36.91 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5404718 113 - 23771897 GGGGGGGGGGAGAGCGGGGAGUGCAAUGUGGAAGUGGCAUGGUG------GGUGGGGCAA-AGUGGCAGCACGUAGCUGCUGCCUGUUGAGGCUGCCUGAUGCCACUUUUGUCUCCGUUU ...........((((((((........(..((((((((((((..------(.(..((((.-.(..(((((.....)))))..).))))..).)..))..))))))))))..))))))))) ( -46.90) >DroSim_CAF1 50550 112 - 1 GGGG-------CAGUGGGGAGUGCAAUGGGAAAGUGGCAUGUUGGCGGGCGGUGGGGCAA-AGUGGCAGCACGUAGCUGCUGCCUGUUGAGGCUGCCUGAUGCCACUUUUGUCUCCGUUU ((((-------((.(....).))).....(((((((((((.....((((((((....(((-...(((((((......)))))))..)))..)))))))))))))))))))...))).... ( -47.10) >DroYak_CAF1 53748 113 - 1 GGGGUGGGGGUGGU-----GUUGGGGCAUGGAAGUGGCAUGAUGG--AGCGGUGGGGCAGCAGUGGCAGCACGUAGCUGCUGCCUGUUGAGGCUGCCUGAUGCCACUUUUGUCUCCGUUU ((((..((((((((-----((..((.(((....))).).......--.(((((....((((((..(((((.....)))))..).)))))..))))))..))))))))))..)).)).... ( -50.50) >consensus GGGG_GGGGG_GAG_GGGGAGUGCAAUGUGGAAGUGGCAUGAUGG___GCGGUGGGGCAA_AGUGGCAGCACGUAGCUGCUGCCUGUUGAGGCUGCCUGAUGCCACUUUUGUCUCCGUUU ((((.........((.......))...(..(((((((((.............(.(((((.....((((((.....))))))((((....))))))))).))))))))))..))))).... (-37.57 = -36.91 + -0.66)

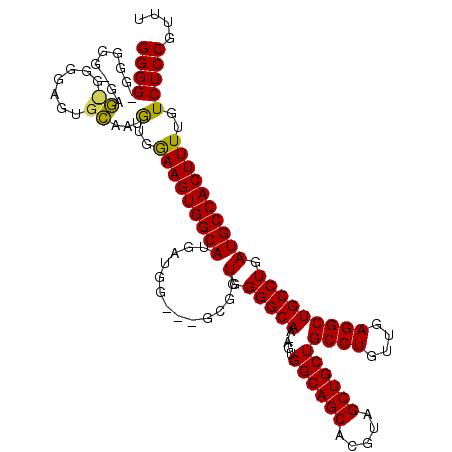
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:57 2006