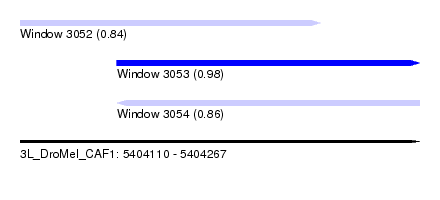
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,404,110 – 5,404,267 |
| Length | 157 |
| Max. P | 0.980182 |

| Location | 5,404,110 – 5,404,228 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -45.78 |
| Consensus MFE | -37.11 |
| Energy contribution | -39.30 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5404110 118 + 23771897 AGA--GAGCCCAAAACUCCGGACCGAACGCAGGUGAAAGCCGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUACUGGCCCAAAAGGGGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCC ...--..((((....((.((..((((((((((((....))).......(..((...))..)..)))(((((((.((.(.(((....))).)))))))))).))))))..)).)).)))). ( -50.30) >DroSec_CAF1 52821 117 + 1 AGA--GAGCCCAAAACUAUGGACCGAACGCAGGUGAAAGCCGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUUCCGGCC-AAAAGGGGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCC ...--..((((....((.((..((((((((((((....))).......(..((...))..)..)))(((((((...((.((-.....)).)).))))))).))))))..)).)).)))). ( -46.60) >DroSim_CAF1 49963 117 + 1 AGAGAGAGCCCAAAACUCCGGACCGAACGCA-GUGAAAGCCGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUACCGGCC-AAAAG-GGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCC .......((((....((.((..(((((((((-(((...((.(....).))....)).)))).....(((((((...((.((-....)-).)).))))))).))))))..)).)).)))). ( -44.70) >DroYak_CAF1 53045 117 + 1 AGA--GAGCCUAAAACUACAACCCAAACGCAGGUGAAAGCCGUGAACCGUGAUACAGGUGCAAUGCUGGUGCCUAGCGGCC-AAAAGGGGCGUGGCAGGAAGUUUGGGGCACGGCAGGCU ...--.(((((..........(((((((((((((....))).((.(((........))).)).)))...((((..(((.((-.....)).)))))))....)))))))((...))))))) ( -41.50) >consensus AGA__GAGCCCAAAACUACGGACCGAACGCAGGUGAAAGCCGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUACCGGCC_AAAAGGGGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCC .......((((....((.((..((((((((((.((...((.(....).))....)).)))).....(((((((...((.((......)).)).))))))).))))))..)).)).)))). (-37.11 = -39.30 + 2.19)


| Location | 5,404,148 – 5,404,267 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -52.12 |
| Consensus MFE | -46.24 |
| Energy contribution | -46.68 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5404148 119 + 23771897 CGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUACUGGCCCAAAAGGGGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCCGUGCCUCGAC-UGCAUGACGCAGAUAACACGGCAACACGG ((((..(((((.((...((((.(((((((((((.((.(.(((....))).))))))))))((((.(((((((.((...))))))))))))-)))))...)))).)).)))))...)))). ( -54.70) >DroSec_CAF1 52859 118 + 1 CGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUUCCGGCC-AAAAGGGGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCCGUGCCUCGAC-UGCAUGACGCAGAUAACACGGCAACACGG ((((..(((((.((...((((.(((((((((((...((.((-.....)).)).)))))))((((.(((((((.((...))))))))))))-)))))...)))).)).)))))...)))). ( -52.60) >DroSim_CAF1 50002 118 + 1 CGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUACCGGCC-AAAAG-GGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCCGUGCCUCGACAUGCAUGAUGCAGAUAACACGGCAACACGG ((((..(((((.((...((((((((((((((((...((.((-....)-).)).))))))).(((.(((((((.((...))))))))))))..))))..))))).)).)))))...)))). ( -52.60) >DroYak_CAF1 53083 118 + 1 CGUGAACCGUGAUACAGGUGCAAUGCUGGUGCCUAGCGGCC-AAAAGGGGCGUGGCAGGAAGUUUGGGGCACGGCAGGCUGUGUCUCGAC-UGCAUGACGCAGAUAACACGGCAACACGG ((((..(((((.((...(((..((((...((((..(((.((-.....)).)))))))...((((.(((((((((....))))))))))))-)))))..)))...)).)))))...)))). ( -48.60) >consensus CGUGAACCGUGAUACAGUUGCAAUGCUCCUGCCUACCGGCC_AAAAGGGGCGUGGCAGGAAGUUUGGGGCGCAGGGGGCCGUGCCUCGAC_UGCAUGACGCAGAUAACACGGCAACACGG ((((..(((((.((...((((.(((((((((((...((.((......)).)).)))))))...(((((((((........)))))))))...))))...)))).)).)))))...)))). (-46.24 = -46.68 + 0.44)


| Location | 5,404,148 – 5,404,267 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -44.33 |
| Consensus MFE | -39.96 |
| Energy contribution | -40.77 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5404148 119 - 23771897 CCGUGUUGCCGUGUUAUCUGCGUCAUGCA-GUCGAGGCACGGCCCCCUGCGCCCCAAACUUCCUGCCACGCCCCUUUUGGGCCAGUAGGCAGGAGCAUUGCAACUGUAUCACGGUUCACG .((((..((((((.(((.((((..((((.-...(.(((.(((....))).))).).....(((((((((((((.....))))..)).)))))))))))))))...))).)))))).)))) ( -49.50) >DroSec_CAF1 52859 118 - 1 CCGUGUUGCCGUGUUAUCUGCGUCAUGCA-GUCGAGGCACGGCCCCCUGCGCCCCAAACUUCCUGCCACGCCCCUUUU-GGCCGGAAGGCAGGAGCAUUGCAACUGUAUCACGGUUCACG .((((..((((((.(((.((((..((((.-...(.(((.(((....))).))).).....(((((((.((((......-)).))...)))))))))))))))...))).)))))).)))) ( -45.70) >DroSim_CAF1 50002 118 - 1 CCGUGUUGCCGUGUUAUCUGCAUCAUGCAUGUCGAGGCACGGCCCCCUGCGCCCCAAACUUCCUGCCACGCC-CUUUU-GGCCGGUAGGCAGGAGCAUUGCAACUGUAUCACGGUUCACG .((((..((((((.(((.((((..((((.....(.(((.(((....))).))).).....(((((((.((.(-(....-)).))...)))))))))))))))...))).)))))).)))) ( -47.40) >DroYak_CAF1 53083 118 - 1 CCGUGUUGCCGUGUUAUCUGCGUCAUGCA-GUCGAGACACAGCCUGCCGUGCCCCAAACUUCCUGCCACGCCCCUUUU-GGCCGCUAGGCACCAGCAUUGCACCUGUAUCACGGUUCACG .((((..((((((.(((..(.((.((((.-((....))...(((((.((.(((.........................-))))).)))))....)))).)).)..))).)))))).)))) ( -34.71) >consensus CCGUGUUGCCGUGUUAUCUGCGUCAUGCA_GUCGAGGCACGGCCCCCUGCGCCCCAAACUUCCUGCCACGCCCCUUUU_GGCCGGUAGGCAGGAGCAUUGCAACUGUAUCACGGUUCACG .((((..((((((.(((.((((..((((.....(.(((.(((....))).))).).....(((((((..(((.......))).....)))))))))))))))...))).)))))).)))) (-39.96 = -40.77 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:55 2006