
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,400,757 – 5,401,020 |
| Length | 263 |
| Max. P | 0.962228 |

| Location | 5,400,757 – 5,400,874 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5400757 117 + 23771897 UCUGCUACAUGGACCCCAUAAUUUGUAAUUUCAACCUAAUUACUUCAAGCCCAGCCGAACGGGGAUCGGA---CCAGCUAAACAAUGUUGUCCGUAAUAAAAUGUCAAAAACCAAAGCAA ..((((...(((.((((.......((((((.......))))))(((..((...)).))).))))..((((---(.(((........)))))))).................))).)))). ( -25.30) >DroSec_CAF1 49600 120 + 1 UCUGCUACAUGGACCCCAUAAUUUGUAAUUUCAACCUAAUUACUUCAAGCCCAGGCGAACGGGGAUAGGAACACCAGCUAAACAAUGUUGUCCGCAAUAAAAUGUCAAAAACCAAAGCAA ..((((...(((.((((.......((((((.......)))))).....((....))....))))...(((....((((........)))))))..................))).)))). ( -25.40) >DroYak_CAF1 49619 117 + 1 UCUGCUACAUGGACCCCAUAAUUUGUAAUUUCAACCUAAUUACUUCAAGCGCAGGCGAAGGGGGCACGGA---CCAGCUAAACAAUGUUGUCCGCAAUAAAAUGUCAAAAACCAAAGCAU ..((((...(((.((((.......((((((.......)))))).....((....))...))))...((((---(.(((........)))))))).................))).)))). ( -27.90) >consensus UCUGCUACAUGGACCCCAUAAUUUGUAAUUUCAACCUAAUUACUUCAAGCCCAGGCGAACGGGGAUCGGA___CCAGCUAAACAAUGUUGUCCGCAAUAAAAUGUCAAAAACCAAAGCAA ..((((...(((.((((.......((((((.......)))))).....((....))....))))...(((....((((........)))))))..................))).)))). (-22.20 = -22.53 + 0.33)

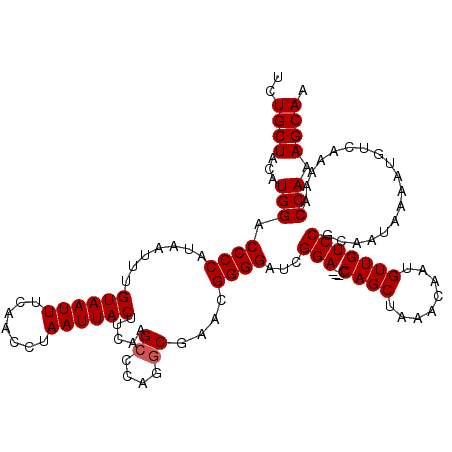

| Location | 5,400,797 – 5,400,905 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5400797 108 + 23771897 UACUUCAAGCCCAGCCGAACGGGGAUCGGA---CCAGCUAAACAAUGUUGUCCGUAAUAAAAUGUCAAAAACCAAAGCAAAACGCCCAUUUCGUCUGAUUUACACA------CACAC--- ...........(((.((((..(((..((((---(.(((........))))))))........(((...........))).....)))..)))).))).........------.....--- ( -20.10) >DroSec_CAF1 49640 117 + 1 UACUUCAAGCCCAGGCGAACGGGGAUAGGAACACCAGCUAAACAAUGUUGUCCGCAAUAAAAUGUCAAAAACCAAAGCAAAACGCCCAUUUCGUCUGAUUCACACACACACACACAC--- ...........((((((((..(((...((....)).(((......(((((....)))))...((........)).)))......)))..))))))))....................--- ( -21.60) >DroYak_CAF1 49659 117 + 1 UACUUCAAGCGCAGGCGAAGGGGGCACGGA---CCAGCUAAACAAUGUUGUCCGCAAUAAAAUGUCAAAAACCAAAGCAUAACGCCCAUUUCGUCUGAAUCACACACACACACACACAAA ...........(((((((((.((((.((((---(.(((........)))))))).......((((...........))))...)))).)))))))))....................... ( -30.90) >consensus UACUUCAAGCCCAGGCGAACGGGGAUCGGA___CCAGCUAAACAAUGUUGUCCGCAAUAAAAUGUCAAAAACCAAAGCAAAACGCCCAUUUCGUCUGAUUCACACACACACACACAC___ ...........((((((((..(((...(((....((((........)))))))((.....................))......)))..))))))))....................... (-19.42 = -19.53 + 0.11)


| Location | 5,400,874 – 5,400,980 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -28.36 |
| Energy contribution | -27.48 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5400874 106 + 23771897 AACGCCCAUUUCGUCUGAUUUACACA------CACAC--------AUGAAGACCAGAAGGACACGCCCCCUCACACGCACAGUGCGUAUGAUUUCGGUUUUUGUGUCCUUUGCUGGUGAC ........((((((.((.........------..)).--------))))))(((((.((((((((((...(((.((((.....)))).)))....)))....)))))))...)))))... ( -29.80) >DroSec_CAF1 49720 112 + 1 AACGCCCAUUUCGUCUGAUUCACACACACACACACAC--------AUGAAGGCCAGAAGGACACGCCCCCUCGCACGCACAGUGCGUAUGAUUUCGGUUUUUGUGUCCUUUGCUGGUGAC ..((((...(((((.((.................)).--------)))))(((..((((((((((((...(((.((((.....)))).)))....)))....)))))))))))))))).. ( -29.83) >DroYak_CAF1 49736 120 + 1 AACGCCCAUUUCGUCUGAAUCACACACACACACACACAAAUACAUAUGAAGGCAAAAAGGAUACGCCCCCUCACACGCACAGGGCGUAUGAUUUUGGUUUUUGUGUCCUUUGCUGGUGAC ..((((...(((((.((.........................)).)))))(((((..((((((((((...(((.((((.....)))).)))....)))....)))))))))))))))).. ( -28.11) >consensus AACGCCCAUUUCGUCUGAUUCACACACACACACACAC________AUGAAGGCCAGAAGGACACGCCCCCUCACACGCACAGUGCGUAUGAUUUCGGUUUUUGUGUCCUUUGCUGGUGAC ..((((......((((.................................))))..((((((((((((...(((.((((.....)))).)))....)))....)))))))))...)))).. (-28.36 = -27.48 + -0.88)

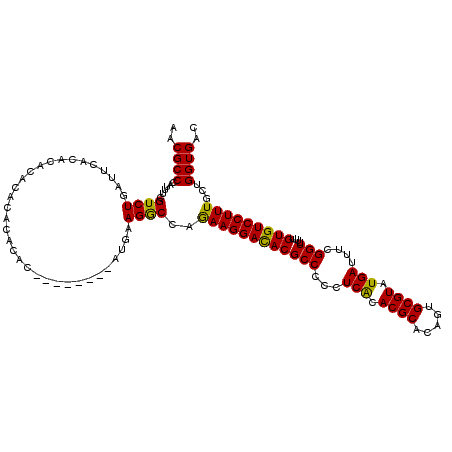

| Location | 5,400,905 – 5,401,020 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -35.39 |
| Energy contribution | -34.07 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5400905 115 + 23771897 -----AUGAAGACCAGAAGGACACGCCCCCUCACACGCACAGUGCGUAUGAUUUCGGUUUUUGUGUCCUUUGCUGGUGACAUAUGUUAAUGAUCUGGCAGCGAUUUCUGGGUCAUUUAAA -----((((...(((((((((((((((...(((.((((.....)))).)))....)))....)))))).((((((.((((((......))).)).).)))))).)))))).))))..... ( -37.90) >DroSec_CAF1 49757 115 + 1 -----AUGAAGGCCAGAAGGACACGCCCCCUCGCACGCACAGUGCGUAUGAUUUCGGUUUUUGUGUCCUUUGCUGGUGACAUAUGUUAAUGAUCUGGCAGCGAUUUCUGGGUCAUUUAAA -----((((...(((((((((((((((...(((.((((.....)))).)))....)))....)))))).((((((.((((((......))).)).).)))))).)))))).))))..... ( -36.10) >DroYak_CAF1 49776 120 + 1 UACAUAUGAAGGCAAAAAGGAUACGCCCCCUCACACGCACAGGGCGUAUGAUUUUGGUUUUUGUGUCCUUUGCUGGUGACAUAUGUAAAUGAUCUGGCAGCGAUUUCUGUGUCAUUUAAA .(((((((..(.((.((((((((((((...(((.((((.....)))).)))....)))....)))))))))..)).)..)))))))(((((((...((((......)))))))))))... ( -36.10) >consensus _____AUGAAGGCCAGAAGGACACGCCCCCUCACACGCACAGUGCGUAUGAUUUCGGUUUUUGUGUCCUUUGCUGGUGACAUAUGUUAAUGAUCUGGCAGCGAUUUCUGGGUCAUUUAAA ......((((((((.((((((((((((...(((.((((.....)))).)))....)))....)))))))))((((.((((((......))).)).).))))........)))).)))).. (-35.39 = -34.07 + -1.32)

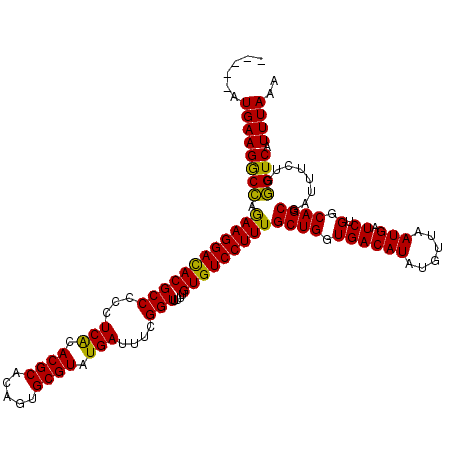
| Location | 5,400,905 – 5,401,020 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -31.79 |
| Energy contribution | -32.90 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5400905 115 - 23771897 UUUAAAUGACCCAGAAAUCGCUGCCAGAUCAUUAACAUAUGUCACCAGCAAAGGACACAAAAACCGAAAUCAUACGCACUGUGCGUGUGAGGGGGCGUGUCCUUCUGGUCUUCAU----- .......((((.((((...((((...((.(((......)))))..))))...((((((.....((....((((((((.....)))))))).))...)))))))))))))).....----- ( -38.20) >DroSec_CAF1 49757 115 - 1 UUUAAAUGACCCAGAAAUCGCUGCCAGAUCAUUAACAUAUGUCACCAGCAAAGGACACAAAAACCGAAAUCAUACGCACUGUGCGUGCGAGGGGGCGUGUCCUUCUGGCCUUCAU----- .....((((.((((((...((((...((.(((......)))))..))))...((((((.....(((....)...(((((.....))))).))....))))))))))))...))))----- ( -33.70) >DroYak_CAF1 49776 120 - 1 UUUAAAUGACACAGAAAUCGCUGCCAGAUCAUUUACAUAUGUCACCAGCAAAGGACACAAAAACCAAAAUCAUACGCCCUGUGCGUGUGAGGGGGCGUAUCCUUUUUGCCUUCAUAUGUA ...(((((((.(((......)))...).))))))(((((((....(((.((((((.((.....((....((((((((.....)))))))).))...)).)))))))))....))))))). ( -32.00) >consensus UUUAAAUGACCCAGAAAUCGCUGCCAGAUCAUUAACAUAUGUCACCAGCAAAGGACACAAAAACCGAAAUCAUACGCACUGUGCGUGUGAGGGGGCGUGUCCUUCUGGCCUUCAU_____ .....((((.((((((...((((...((.(((......)))))..))))...((((((.....((....((((((((.....)))))))).))...))))))))))))...))))..... (-31.79 = -32.90 + 1.11)

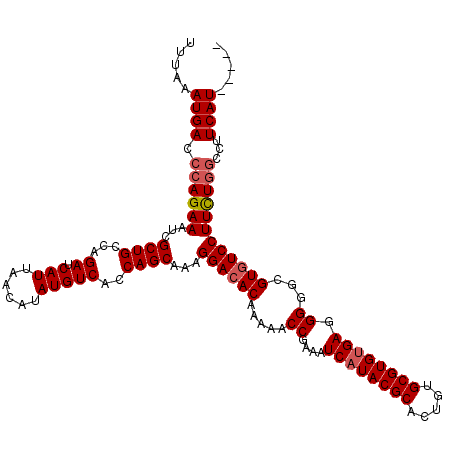
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:49 2006