
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,398,607 – 5,398,804 |
| Length | 197 |
| Max. P | 0.924087 |

| Location | 5,398,607 – 5,398,725 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -31.43 |
| Energy contribution | -31.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5398607 118 - 23771897 GACUUGUGCUGGUGGGAGCAGCAGCAGGGAGAAUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCAGGAGCCAUGGGGC--AGGAUCCAGAUCCUGCAGGAGCAAA ..(((.(((((.((....)).))))).).))....((((...................(((((......))))).((((((((....((((..--....))))..))))))))..)))). ( -37.20) >DroSec_CAF1 47511 112 - 1 GACUUGUGCUGG---G---AGCAGCAGGGAGAAUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCAGGAGACAUGGGGC--AGGAUCCAGAUCCUGCAGGAGCAAG ..(((((((((.---.---(((((((.......))))))...................((....))...)..))))(((((((....((((..--....))))..)))))))...))))) ( -36.10) >DroYak_CAF1 47453 114 - 1 GACUUGUGCUGG---A---AGCAGCAGGGAGAGUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCAGGAGCCAUGGGGCACAGGAUCCAGAUCCUGCUGGAGCAAA ...((.(((((.---.---..))))).)).(((((((((........................)))))))))..(((.(((..(((....))).((((((....))))))))).)))... ( -35.76) >consensus GACUUGUGCUGG___G___AGCAGCAGGGAGAAUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCAGGAGCCAUGGGGC__AGGAUCCAGAUCCUGCAGGAGCAAA ...((((((((.........((((((.......)))))).............(((((.((....)).))))))))).((((((....((((........))))..))))))....)))). (-31.43 = -31.43 + 0.00)

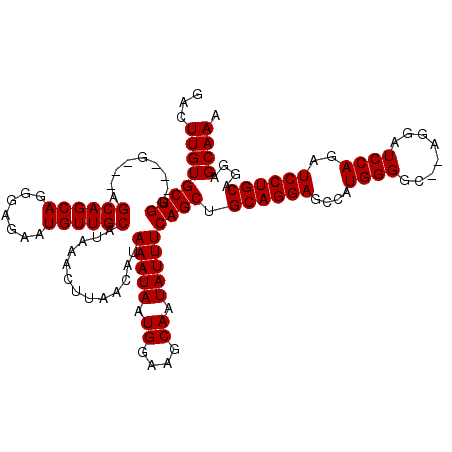

| Location | 5,398,645 – 5,398,765 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -24.01 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5398645 120 - 23771897 GAUAUUAAAAACAAGAGAGGAAGGACUUGUGCUGCAGCAGGACUUGUGCUGGUGGGAGCAGCAGCAGGGAGAAUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCA ..........(((((..........)))))...(((((.....((.(((((.((....)).))))).)).(((((((((........................)))))))))..))))). ( -29.86) >DroSec_CAF1 47549 114 - 1 GAUAUUAAAAACAAGAGAGGAAGGACUUGUGCUGCAGCAGGACUUGUGCUGG---G---AGCAGCAGGGAGAAUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCA ..........(((((..........)))))...(((((.....((.(((((.---.---..))))).)).(((((((((........................)))))))))..))))). ( -26.76) >DroYak_CAF1 47493 114 - 1 GAUAUUAAAAACAAGAGAGAAAGGACUAGUGCAGCAGCAGGACUUGUGCUGG---A---AGCAGCAGGGAGAGUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCA .............................((((((.((((.((((.(((((.---.---..)))))....)))).))))...................(((((......))))))))))) ( -29.60) >consensus GAUAUUAAAAACAAGAGAGGAAGGACUUGUGCUGCAGCAGGACUUGUGCUGG___G___AGCAGCAGGGAGAAUGUUGCAUAAACUUAACAUAAAUAAUGGAAGCAAUAUUUCAGCUGCA ..........(((((..........)))))...(((((....((((((((.........)))).))))..(((((((((........................)))))))))..))))). (-24.01 = -24.13 + 0.11)

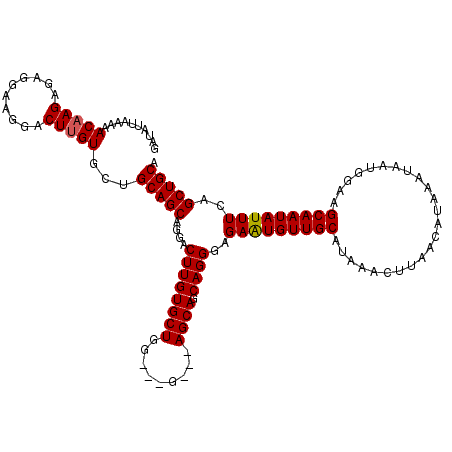
| Location | 5,398,685 – 5,398,804 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -19.73 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5398685 119 + 23771897 UGCAACAUUCUCCCUGCUGCUGCUCCCACCAGCACAAGUCCUGCUGCAGCACAAGUCCUUCCUCUCUUGUUUUUAAUAUCCUUCAGCUUGUAAAGGAAUUACUCCUCAAGCCGGCA-ACU (((............(((((((((......))))..((.....)))))))(((((..........)))))...............(((((...((((.....)))))))))..)))-... ( -24.10) >DroSec_CAF1 47589 113 + 1 UGCAACAUUCUCCCUGCUGCU---C---CCAGCACAAGUCCUGCUGCAGCACAAGUCCUUCCUCUCUUGUUUUUAAUAUCCUUCAGCUUGUAAAGGAAUUACUCCUCAAGCCGGCA-ACU (((............(((((.---.---..((((.......)))))))))(((((..........)))))...............(((((...((((.....)))))))))..)))-... ( -23.70) >DroYak_CAF1 47533 114 + 1 UGCAACACUCUCCCUGCUGCU---U---CCAGCACAAGUCCUGCUGCUGCACUAGUCCUUUCUCUCUUGUUUUUAAUAUCCUUCAGCUUGUAAAGGAAUUACUCCUCAAGCCGGCAAACU (((..........(((.(((.---.---.(((((.......)))))..))).)))..............................(((((...((((.....)))))))))..))).... ( -21.60) >consensus UGCAACAUUCUCCCUGCUGCU___C___CCAGCACAAGUCCUGCUGCAGCACAAGUCCUUCCUCUCUUGUUUUUAAUAUCCUUCAGCUUGUAAAGGAAUUACUCCUCAAGCCGGCA_ACU .(((.((..((...(((((..........)))))..))...)).))).(((((((..........)))))...............(((((...((((.....)))))))))..))..... (-19.73 = -20.07 + 0.33)

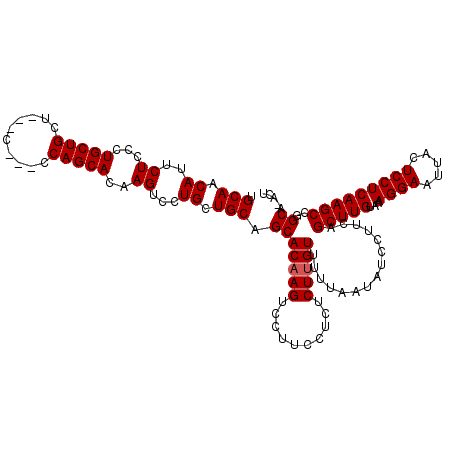
| Location | 5,398,685 – 5,398,804 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -29.23 |
| Energy contribution | -29.57 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5398685 119 - 23771897 AGU-UGCCGGCUUGAGGAGUAAUUCCUUUACAAGCUGAAGGAUAUUAAAAACAAGAGAGGAAGGACUUGUGCUGCAGCAGGACUUGUGCUGGUGGGAGCAGCAGCAGGGAGAAUGUUGCA ...-.(((((((((..(((......)))..))))))).............(((((..........))))))).((((((....((.(((((.((....)).))))).))....)))))). ( -36.10) >DroSec_CAF1 47589 113 - 1 AGU-UGCCGGCUUGAGGAGUAAUUCCUUUACAAGCUGAAGGAUAUUAAAAACAAGAGAGGAAGGACUUGUGCUGCAGCAGGACUUGUGCUGG---G---AGCAGCAGGGAGAAUGUUGCA ...-.(((((((((..(((......)))..))))))).............(((((..........))))))).((((((....((.(((((.---.---..))))).))....)))))). ( -33.00) >DroYak_CAF1 47533 114 - 1 AGUUUGCCGGCUUGAGGAGUAAUUCCUUUACAAGCUGAAGGAUAUUAAAAACAAGAGAGAAAGGACUAGUGCAGCAGCAGGACUUGUGCUGG---A---AGCAGCAGGGAGAGUGUUGCA ....((((((((((..(((......)))..)))))))..............((..((........))..))..)))((((.((((.(((((.---.---..)))))....)))).)))). ( -32.00) >consensus AGU_UGCCGGCUUGAGGAGUAAUUCCUUUACAAGCUGAAGGAUAUUAAAAACAAGAGAGGAAGGACUUGUGCUGCAGCAGGACUUGUGCUGG___G___AGCAGCAGGGAGAAUGUUGCA .....(((((((((..(((......)))..))))))).............(((((..........))))))).((((((...((((((((.........)))).)))).....)))))). (-29.23 = -29.57 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:41 2006