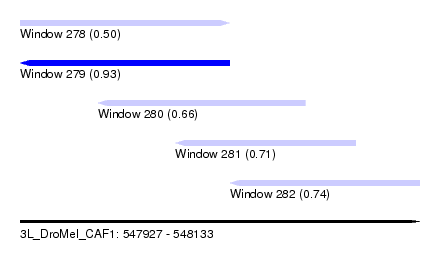
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 547,927 – 548,133 |
| Length | 206 |
| Max. P | 0.928608 |

| Location | 547,927 – 548,035 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 547927 108 + 23771897 GUUGCCGCCGUUGGUGCAUUUGAUUUAUUGCAUUUUCCAUUUGGCUAAGCGUGGAAAAUGCGAAAAUACGAACUUCCGAUUGCCAUUUGGAAAAUA--------UGUAAAUG----GCGU .....(((((((..((((((((((((.(((((((((((((...((...))))))))))))))))))).)))).((((((.......))))))....--------))))))))----))). ( -33.90) >DroEre_CAF1 25779 108 + 1 GUUGCCGCCGUUGGUGCAUUUGAUUUAUUGCAUUUUCCAUUUGGCUAAGCGUGGAAAAUGCGAAAAUACGAACUUCCGAUUGCCAUUUGGAAAAUA--------UGUAAAUG----GCGU .....(((((((..((((((((((((.(((((((((((((...((...))))))))))))))))))).)))).((((((.......))))))....--------))))))))----))). ( -33.90) >DroYak_CAF1 22597 116 + 1 GUUGCCGCCGCUGGUGCAUUUGAUUUAUUGCAUUUUCCAUUUGGCUAAGCGUGGCAAAUGCGAAAAUACGAACUUCCGAUUGCCAUUUGGAAAAUAUGUGCAUAUGUAAAUG----GCGU .....((((((....)).....(((((((((((((((((..((((...((((.....)))).......((......))...))))..))))))....)))))...)))))))----))). ( -33.70) >DroAna_CAF1 29101 109 + 1 GUUGUUG-----------UAUGAUUUAUUGCAUUUUCCAUUUGGCGGAGCGUGGAAAAUGUGAAAAUCCCAACUUCCGAUUGCCAUUUUGAAGAGAGAGAGAGAGAGAGAUGUGGUGUGU ...((((-----------...(((((.(..((((((((((...((...))))))))))))..)))))).))))........(((((.........................))))).... ( -22.11) >consensus GUUGCCGCCGUUGGUGCAUUUGAUUUAUUGCAUUUUCCAUUUGGCUAAGCGUGGAAAAUGCGAAAAUACGAACUUCCGAUUGCCAUUUGGAAAAUA________UGUAAAUG____GCGU .....(((..........((((((((.(((((((((((((...((...))))))))))))))))))).)))).((((((.......))))))........................))). (-19.19 = -19.88 + 0.69)

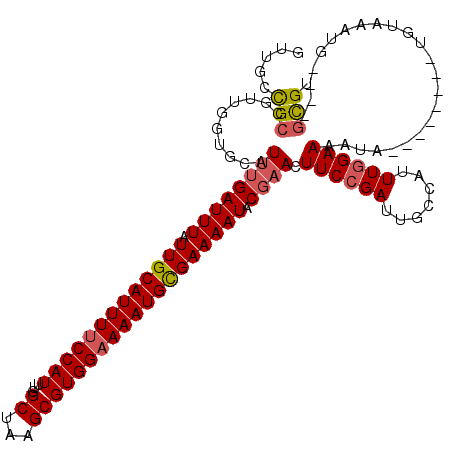
| Location | 547,927 – 548,035 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -14.18 |
| Energy contribution | -15.18 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 547927 108 - 23771897 ACGC----CAUUUACA--------UAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUUCCACGCUUAGCCAAAUGGAAAAUGCAAUAAAUCAAAUGCACCAACGGCGGCAAC .(((----(.......--------.((((((((..((((....((((((....))))))((.........))...))))..))))))))(((..........)))......))))..... ( -27.50) >DroEre_CAF1 25779 108 - 1 ACGC----CAUUUACA--------UAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUUCCACGCUUAGCCAAAUGGAAAAUGCAAUAAAUCAAAUGCACCAACGGCGGCAAC .(((----(.......--------.((((((((..((((....((((((....))))))((.........))...))))..))))))))(((..........)))......))))..... ( -27.50) >DroYak_CAF1 22597 116 - 1 ACGC----CAUUUACAUAUGCACAUAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUGCCACGCUUAGCCAAAUGGAAAAUGCAAUAAAUCAAAUGCACCAGCGGCGGCAAC .(((----(.........((((....(((((((..((((((.(((((((....)))))))...)))))).((...))....)))))))))))...........((....))))))..... ( -30.90) >DroAna_CAF1 29101 109 - 1 ACACACCACAUCUCUCUCUCUCUCUCUCUUCAAAAUGGCAAUCGGAAGUUGGGAUUUUCACAUUUUCCACGCUCCGCCAAAUGGAAAAUGCAAUAAAUCAUA-----------CAACAAC ...............................................((((.(((((...(((((((((.((...))....)))))))))....)))))...-----------))))... ( -16.60) >consensus ACGC____CAUUUACA________UAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUUCCACGCUUAGCCAAAUGGAAAAUGCAAUAAAUCAAAUGCACCAACGGCGGCAAC ........................(((((((((..((((....((((((....))))))((.........))...))))..))))))))).............................. (-14.18 = -15.18 + 1.00)


| Location | 547,967 – 548,074 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 547967 107 - 23771897 -CGGCAACAACGGGCGCUGCAUGCAACAUGUUGCCGAGAAACGC----CAUUUACA--------UAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUUCCACGCUUAGCCAA -((((((((...(..((.....))..).))))))))......((----(((((...--------........)))))))....(((((..((......))...)))))............ ( -27.70) >DroEre_CAF1 25819 107 - 1 -UGGCAACAACUGGCGUUGCAUGCAACAUGUUGCCAAGAAACGC----CAUUUACA--------UAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUUCCACGCUUAGCCAA -((((......(((((..(((((...))))))))))......((----(((((...--------........)))))))....(((((..((......))...))))).......)))). ( -27.70) >DroYak_CAF1 22637 115 - 1 -CGGCAACAACGGGCGUUGCAUGCAACAUGUUGCCAAGAAACGC----CAUUUACAUAUGCACAUAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUGCCACGCUUAGCCAA -.(((.......((((((....((((....)))).....)))))----)..........................((((((.(((((((....)))))))...))))))......))).. ( -32.40) >DroAna_CAF1 29130 120 - 1 UAGCCAUUAGACGGCAUUGCAUGCAACAUGUUGCCACCAAACACACCACAUCUCUCUCUCUCUCUCUCUUCAAAAUGGCAAUCGGAAGUUGGGAUUUUCACAUUUUCCACGCUCCGCCAA ..((((((....((((..(((((...))))))))).............................(......).))))))...((((.(((((((..........))))).)))))).... ( -21.80) >consensus _CGGCAACAACGGGCGUUGCAUGCAACAUGUUGCCAAGAAACGC____CAUUUACA________UAUUUUCCAAAUGGCAAUCGGAAGUUCGUAUUUUCGCAUUUUCCACGCUUAGCCAA ..(((((((....(((.....)))....)))))))........................................((((....((((((....))))))((.........))...)))). (-19.00 = -19.38 + 0.38)

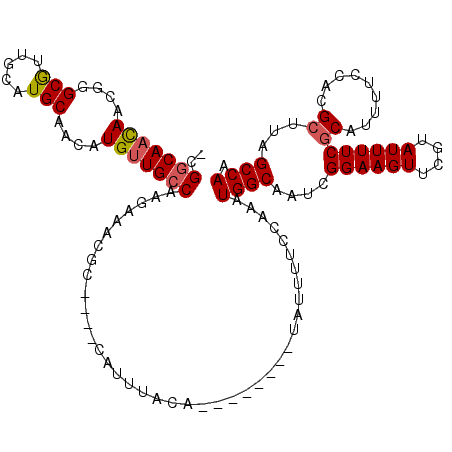
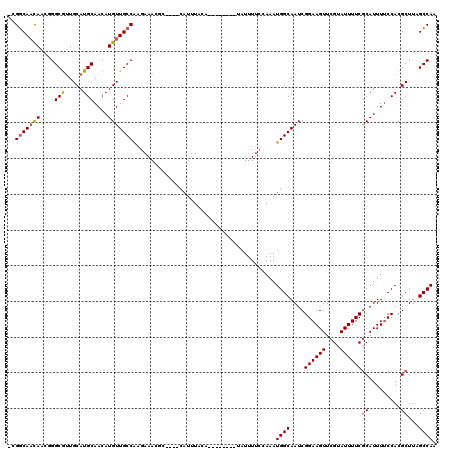
| Location | 548,007 – 548,100 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.92 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 548007 93 - 23771897 GGC------UAAUGAUGAUUGA-----AUCGAUGGCC----CGGCAACAACGGGCGCUGCAUGCAACAUGUUGCCGAGAAACGC----CAUUUACA--------UAUUUUCCAAAUGGCA (((------((.((((......-----)))).)))))----((((((((...(..((.....))..).))))))))......((----(((((...--------........))))))). ( -32.40) >DroEre_CAF1 25859 93 - 1 GGU------UAAUGAUGAUUGA-----AUCGAUGGCC----UGGCAACAACUGGCGUUGCAUGCAACAUGUUGCCAAGAAACGC----CAUUUACA--------UAUUUUCCAAAUGGCA (((------((.((((......-----)))).)))))----((((((((....((((...))))....))))))))......((----(((((...--------........))))))). ( -29.70) >DroYak_CAF1 22677 107 - 1 GGAUGAUGAUAAUGAUGAUUGA-----AUCGAUGGCC----CGGCAACAACGGGCGUUGCAUGCAACAUGUUGCCAAGAAACGC----CAUUUACAUAUGCACAUAUUUUCCAAAUGGCA ...((..((((.((((......-----))))...(((----((.......)))))((((....)))).))))..))......((----(((((..((((....)))).....))))))). ( -25.50) >DroAna_CAF1 29170 114 - 1 GUU------GAACGAUGACUAGCCAUUAUGACUAGCAUAUUAGCCAUUAGACGGCAUUGCAUGCAACAUGUUGCCACCAAACACACCACAUCUCUCUCUCUCUCUCUCUUCAAAAUGGCA ...------............((((((.((((((((......))...)))..((((..(((((...)))))))))..................................))).)))))). ( -21.00) >consensus GGU______UAAUGAUGAUUGA_____AUCGAUGGCC____CGGCAACAACGGGCGUUGCAUGCAACAUGUUGCCAAGAAACGC____CAUUUACA________UAUUUUCCAAAUGGCA .............(((((((((......))))).........(((((((....(((.....)))....))))))).............))))............................ (-12.61 = -12.67 + 0.06)

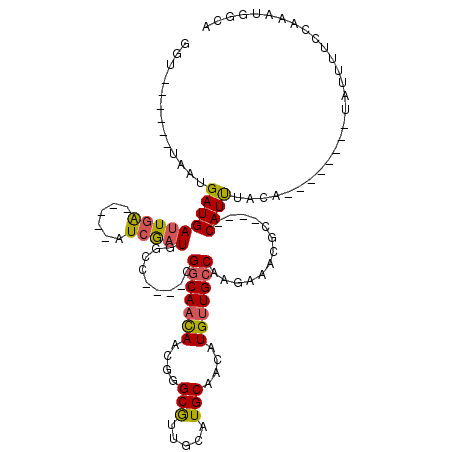
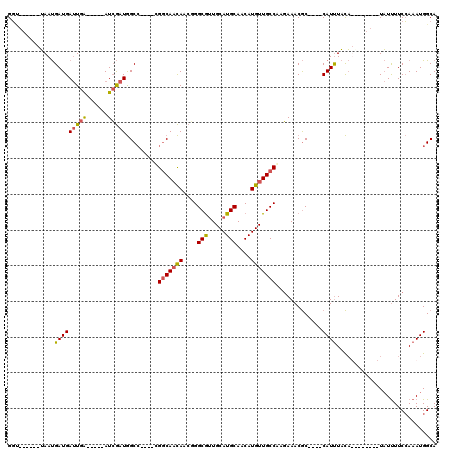
| Location | 548,035 – 548,133 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.80 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -17.55 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 548035 98 - 23771897 GCAUCGUUAGCUU--CAGCUGG--ACAUGGCAUCGAU---GGC------UAAUGAUGAUUGA-----AUCGAUGGCC----CGGCAACAACGGGCGCUGCAUGCAACAUGUUGCCGAGAA .((((((((((((--(.(((..--....)))...)).---)))------)))))))).....-----.(((...(((----((.......)))))((.(((((...))))).)))))... ( -35.10) >DroEre_CAF1 25887 98 - 1 GCAUCGCUAGCUC--CAGCUGG--ACAUGGCACCGAU---GGU------UAAUGAUGAUUGA-----AUCGAUGGCC----UGGCAACAACUGGCGUUGCAUGCAACAUGUUGCCAAGAA ((....(((((..--..)))))--.....))......---(((------((.((((......-----)))).)))))----((((((((....((((...))))....)))))))).... ( -31.40) >DroYak_CAF1 22713 108 - 1 GCAUCGUUAGCUCUACAGCUGGCAACAUGGCAUCGAU---GGAUGAUGAUAAUGAUGAUUGA-----AUCGAUGGCC----CGGCAACAACGGGCGUUGCAUGCAACAUGUUGCCAAGAA ...((...((((....))))((((((((..(((((((---.(((.((.......)).)))..-----)))))))(((----((.......)))))((((....))))))))))))..)). ( -39.10) >DroAna_CAF1 29210 105 - 1 CGAUCGUUAAUAC--CAGCU-------UAGCCGCGAUAAUGUU------GAACGAUGACUAGCCAUUAUGACUAGCAUAUUAGCCAUUAGACGGCAUUGCAUGCAACAUGUUGCCACCAA .............--..((.-------..((((...(((((((------((...(((.((((.(.....).))))))).)))).)))))..))))...))..((((....))))...... ( -22.40) >consensus GCAUCGUUAGCUC__CAGCUGG__ACAUGGCAUCGAU___GGU______UAAUGAUGAUUGA_____AUCGAUGGCC____CGGCAACAACGGGCGUUGCAUGCAACAUGUUGCCAAGAA .((((((((........(((........))).((......)).......)))))))).........................(((((((....(((.....)))....)))))))..... (-17.55 = -17.55 + 0.00)

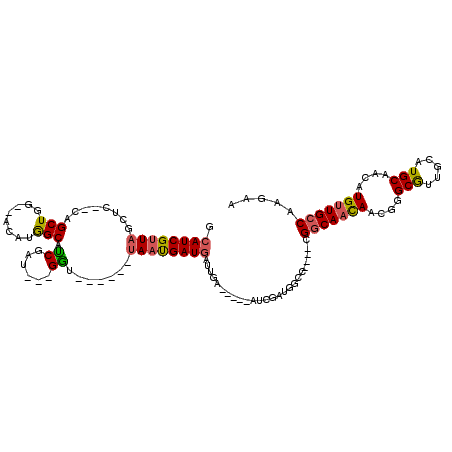
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:29 2006