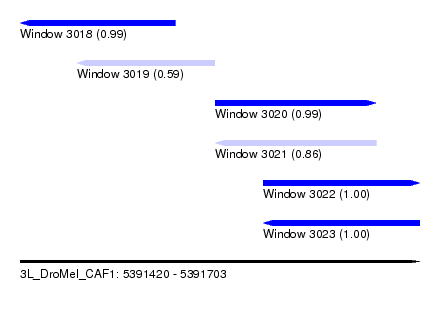
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,391,420 – 5,391,703 |
| Length | 283 |
| Max. P | 0.999976 |

| Location | 5,391,420 – 5,391,530 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -21.82 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5391420 110 - 23771897 GAAAAUACGUUUUUAAUUUCAUUGUUUACUCUGCUCCACAGGAGCUAAACAAGGAGAUUUAUGAACGUAAACUAAACUUUCGCAUAGUCUAAGGAUCAGCCUCAUAAAAA .....(((((((..((((((.(((((((....(((((...)))))))))))).))))))...)))))))............((...(((....)))..)).......... ( -25.00) >DroSec_CAF1 40436 110 - 1 GAAAAUAUGUUUUUAAUUUCAUUGUUUGCUCUGCCCCACAAGAGCUAAACAAGGAGAUUUAUGAACGUAAAGGAAACUUUGGCAUAGUCUAAGGAUCAUCCUCAUAAAAA .....(((((((..((((((.(((((((((((........))))).)))))).))))))...))))))).((((..((((((......))))))....))))........ ( -28.90) >DroSim_CAF1 40079 110 - 1 GAAAAUAUGUUUUUAAUUUCAUUGUUUGUUCUGCCCCAUAAGAGCUAAACAAAGAGAUUUAUGAACGUAAAGGAAACUUUGGCAUAGUCUAAGGAUCAUCCUCAUAAAAA .....(((((((..((((((.(((((((((((........))))).)))))).))))))...))))))).((((..((((((......))))))....))))........ ( -26.50) >DroEre_CAF1 40613 110 - 1 GAAAAUAUGUUUUUAAUUUCAUUGUCUGCUCCGCCCCACAAGAGCUAAACAAGGAGAUUUAUGGGCGCAAAGGAAACUUUCGCAUAUGCCAAGGAUGAGCCUCAUAAAAU ................((((.((((..((((..........))))...)))).))))(((((((((((((((....)))).)).....(....)....))).)))))).. ( -25.30) >DroYak_CAF1 40149 110 - 1 GAAAAUAUGUUUUUAAUUUCAUUGUUUGCUCUGCCCCACAAGAGCUAAAGAAGGAGAUUUAUGAGCGUAAAGGAAACUUUCGCAUAUGCCAAGGAUGAGCCUCAUAAAAA ................((((.((.((((((((........))))).))).)).))))((((((((((.((((....)))))))....((((....)).)).))))))).. ( -26.40) >consensus GAAAAUAUGUUUUUAAUUUCAUUGUUUGCUCUGCCCCACAAGAGCUAAACAAGGAGAUUUAUGAACGUAAAGGAAACUUUCGCAUAGUCUAAGGAUCAGCCUCAUAAAAA ........((((..((((((.(((((((((((........))))).)))))).))))))...))))((((((....)))).))........(((.....)))........ (-21.82 = -21.98 + 0.16)



| Location | 5,391,460 – 5,391,558 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5391460 98 - 23771897 GAAUCUAAAUCCCGACUGUGCCCGACC---------AGAAAAUACGUUUUUAAUUUCAUUGUUUACUCUGCUCCACAGGAGCUAAACAAGGAGAUUUAUGAACGUAA ...............(((........)---------))....(((((((..((((((.(((((((....(((((...)))))))))))).))))))...))))))). ( -24.70) >DroSec_CAF1 40476 107 - 1 GAAUCUAAAUCCCGACAGUGCCCGACCAGUAUGAGCAGAAAAUAUGUUUUUAAUUUCAUUGUUUGCUCUGCCCCACAAGAGCUAAACAAGGAGAUUUAUGAACGUAA .............(...((((.......))))...)......(((((((..((((((.(((((((((((........))))).)))))).))))))...))))))). ( -24.60) >DroSim_CAF1 40119 98 - 1 GAAUCUAAAUCCCGACUGUGCCCGACC---------AGAAAAUAUGUUUUUAAUUUCAUUGUUUGUUCUGCCCCAUAAGAGCUAAACAAAGAGAUUUAUGAACGUAA ...............(((........)---------))....(((((((..((((((.(((((((((((........))))).)))))).))))))...))))))). ( -21.90) >DroEre_CAF1 40653 98 - 1 GAAUCUAAAUCCCGACUUGGGCCGUCC---------AGAAAAUAUGUUUUUAAUUUCAUUGUCUGCUCCGCCCCACAAGAGCUAAACAAGGAGAUUUAUGGGCGCAA ..........(((.....))).(((((---------((((((....)))))((((((.((((..((((..........))))...)))).))))))..))))))... ( -20.90) >DroYak_CAF1 40189 98 - 1 GAAUCUAAAUCCCGCCAGUGCCCGUCC---------AGAAAAUAUGUUUUUAAUUUCAUUGUUUGCUCUGCCCCACAAGAGCUAAAGAAGGAGAUUUAUGAGCGUAA ...........................---------......(((((((..((((((.((.((((((((........))))).))).)).))))))...))))))). ( -19.20) >consensus GAAUCUAAAUCCCGACUGUGCCCGACC_________AGAAAAUAUGUUUUUAAUUUCAUUGUUUGCUCUGCCCCACAAGAGCUAAACAAGGAGAUUUAUGAACGUAA ..........................................(((((((..((((((.(((((((((((........))))).)))))).))))))...))))))). (-18.96 = -19.12 + 0.16)


| Location | 5,391,558 – 5,391,672 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5391558 114 + 23771897 AAUUAAACUGUCAACGGGACA------GGGUCUUCCAUUCACGGCAAACAAGCCGCCCUCCUCCGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGU ...(((((((.(((.(((.((------(((...........((((......))))............)))))))).((((((((((......)))).))))))....))).)).))))). ( -35.30) >DroSec_CAF1 40583 114 + 1 AAUUAAACUGUCAACGGGACA------GGGACUCCCAUUCACCGCAAACAAGUCGCACUCCUACGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGU .......(((((.....))))------)((....)).......(((((((....(((.......((.....))...((((((((((......)))).)))))).....)))..))))))) ( -32.50) >DroSim_CAF1 40217 114 + 1 AAUUAAACUGUCAACGGGACA------GGGACUCCCAUUCACCGCAAACAAGCCGCACUCCUCCGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGU .......(((((.....))))------)((....)).......(((((((....(((.......((.....))...((((((((((......)))).)))))).....)))..))))))) ( -32.50) >DroEre_CAF1 40751 120 + 1 AAUUAAACUGUCAACGGGACCGGGACUGGGACUCCCAUUCACCGCAAACAAGCCGCACUCCUCCGCCUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGU ...(((((((.((((((((.(((((.((((...)))).)).)))..........((........)).)))))....((((((((((......)))).))))))....))).)).))))). ( -35.40) >DroYak_CAF1 40287 113 + 1 AAUUAAACUGUCAACGGGACA-------GGACUCCCAUUCACCGCAAACCAGCCGCACUCCUUCGUUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGU ...(((((((.(((.(((.((-------(((............((......)).((........)).)))))))).((((((((((......)))).))))))....))).)).))))). ( -29.40) >consensus AAUUAAACUGUCAACGGGACA______GGGACUCCCAUUCACCGCAAACAAGCCGCACUCCUCCGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGU ...............((((.............)))).......(((((((....(((.......((.....))...((((((((((......)))).)))))).....)))..))))))) (-27.30 = -27.38 + 0.08)


| Location | 5,391,558 – 5,391,672 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -40.94 |
| Consensus MFE | -35.62 |
| Energy contribution | -36.42 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5391558 114 - 23771897 ACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGGAGGAGGGCGGCUUGUUUGCCGUGAAUGGAAGACCC------UGUCCCGUUGACAGUUUAAUU .((..((((((......((((((.((((......)))))))))).(((((((..((........))..)))))))))))))..)).......(------((((.....)))))....... ( -40.80) >DroSec_CAF1 40583 114 - 1 ACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGUAGGAGUGCGACUUGUUUGCGGUGAAUGGGAGUCCC------UGUCCCGUUGACAGUUUAAUU ..((((...((......((((((.((((......)))))))))).(((((((..(((......)))..))))))))).((.(((((((.....------..))))))).)).)))).... ( -40.80) >DroSim_CAF1 40217 114 - 1 ACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGGAGGAGUGCGGCUUGUUUGCGGUGAAUGGGAGUCCC------UGUCCCGUUGACAGUUUAAUU ..((((...((......((((((.((((......)))))))))).(((((((..(((......)))..))))))))).((.(((((((.....------..))))))).)).)))).... ( -41.00) >DroEre_CAF1 40751 120 - 1 ACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAGGCGGAGGAGUGCGGCUUGUUUGCGGUGAAUGGGAGUCCCAGUCCCGGUCCCGUUGACAGUUUAAUU ..((((.(((((..(.....)..))))).((.(((((.(((..(..((((((..(((......)))..))))))..)((.((.((((...)))).)))).)))))))).)).)))).... ( -42.60) >DroYak_CAF1 40287 113 - 1 ACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAACGAAGGAGUGCGGCUGGUUUGCGGUGAAUGGGAGUCC-------UGUCCCGUUGACAGUUUAAUU .(((((.((((..((..((((((.((((......))))))))))...)).(....)...........)))))))))..((.(((((((....-------..))))))).))......... ( -39.50) >consensus ACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGGAGGAGUGCGGCUUGUUUGCGGUGAAUGGGAGUCCC______UGUCCCGUUGACAGUUUAAUU ..((((...((......((((((.((((......)))))))))).(((((((..(((......)))..))))))))).((.(((((((.............))))))).)).)))).... (-35.62 = -36.42 + 0.80)



| Location | 5,391,592 – 5,391,703 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -35.54 |
| Energy contribution | -35.06 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.14 |
| SVM RNA-class probability | 0.999976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5391592 111 + 23771897 ACGGCAAACAAGCCGCCCUCCUCCGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGUCAUGUUGAGCAGCAGGCAUUAAAUUCGAAUU .((((......)))).........((((.(((((((((((((((((......)))).))))))........((((.....)))).))))))).)))).............. ( -41.00) >DroSec_CAF1 40617 111 + 1 ACCGCAAACAAGUCGCACUCCUACGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGUCAUGUUGAGCAGCAGGCAUUAAAUUCGAAUU ...((.........))........((((.(((((((((((((((((......)))).))))))........((((.....)))).))))))).)))).............. ( -35.40) >DroSim_CAF1 40251 111 + 1 ACCGCAAACAAGCCGCACUCCUCCGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGUCAUGUUGAGCAGCAGGCAUUAAAUUCGAAUU ...((......))...........((((.(((((((((((((((((......)))).))))))........((((.....)))).))))))).)))).............. ( -35.50) >DroEre_CAF1 40791 111 + 1 ACCGCAAACAAGCCGCACUCCUCCGCCUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGUCAUGUUGAGCAGAAGGCAUUAAAUUCGAAUU ...((......))...........((((.(((((((((((((((((......)))).))))))........((((.....)))).))))))).)))).............. ( -38.20) >DroYak_CAF1 40320 111 + 1 ACCGCAAACCAGCCGCACUCCUUCGUUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGUCAUGUUGAGCAGAAGGCAUUAAAUUCGAAUU ...........(((((........))...(((((((((((((((((......)))).))))))........((((.....)))).)))))))..))).............. ( -33.50) >consensus ACCGCAAACAAGCCGCACUCCUCCGCUUCCUGCUCACCUGUCGCCAUUCACCUGGCAGACAGGACACUUGCCAUGUUUGUCAUGUUGAGCAGCAGGCAUUAAAUUCGAAUU ...((......))...........((((.(((((((((((((((((......)))).))))))........((((.....)))).))))))).)))).............. (-35.54 = -35.06 + -0.48)

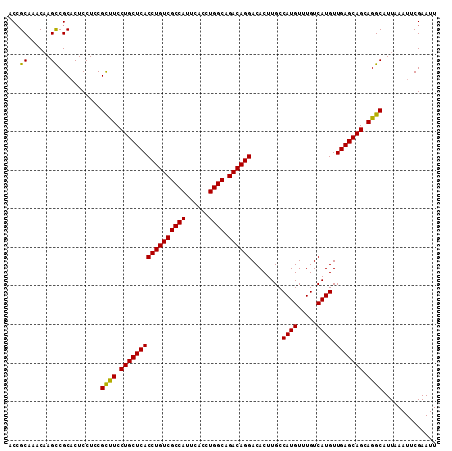
| Location | 5,391,592 – 5,391,703 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -42.04 |
| Consensus MFE | -37.06 |
| Energy contribution | -37.74 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5391592 111 - 23771897 AAUUCGAAUUUAAUGCCUGCUGCUCAACAUGACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGGAGGAGGGCGGCUUGUUUGCCGU ........(((..(((.(.(((((((.((((.....))))........((((((.((((......))))))))))))))))).).)))...))).(((((......))))) ( -42.40) >DroSec_CAF1 40617 111 - 1 AAUUCGAAUUUAAUGCCUGCUGCUCAACAUGACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGUAGGAGUGCGACUUGUUUGCGGU ...(((..((..((((.(.(((((((.((((.....))))........((((((.((((......))))))))))))))))).).))))..))...)))............ ( -39.20) >DroSim_CAF1 40251 111 - 1 AAUUCGAAUUUAAUGCCUGCUGCUCAACAUGACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGGAGGAGUGCGGCUUGUUUGCGGU ..............(((..(((((((.((((.....))))........((((((.((((......)))))))))))))))))...(((((.(((.....))).)))))))) ( -41.40) >DroEre_CAF1 40791 111 - 1 AAUUCGAAUUUAAUGCCUUCUGCUCAACAUGACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAGGCGGAGGAGUGCGGCUUGUUUGCGGU ....((.((((..(((((((((((((.((((.....))))........((((((.((((......)))))))))))))))))))))))...)))).))((......))... ( -47.80) >DroYak_CAF1 40320 111 - 1 AAUUCGAAUUUAAUGCCUUCUGCUCAACAUGACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAACGAAGGAGUGCGGCUGGUUUGCGGU ....((((((...(((((((((((((.((((.....))))........((((((.((((......))))))))))))))).(....)..))))).)))...)))))).... ( -39.40) >consensus AAUUCGAAUUUAAUGCCUGCUGCUCAACAUGACAAACAUGGCAAGUGUCCUGUCUGCCAGGUGAAUGGCGACAGGUGAGCAGGAAGCGGAGGAGUGCGGCUUGUUUGCGGU ..............(((..(((((((.((((.....))))........((((((.((((......)))))))))))))))))...(((((.(((.....))).)))))))) (-37.06 = -37.74 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:26 2006