
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,386,168 – 5,386,297 |
| Length | 129 |
| Max. P | 0.995019 |

| Location | 5,386,168 – 5,386,261 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5386168 93 + 23771897 AAACAACACA--AACACAACAAGUACCCAGCUCAAUUUUGUGUUCUUUUUGG-GGAAA-AACUUUUCGCCAGCAAAGACAAAACACCUUGGGGGUGC ..........--..........((((((...((((....(((((((((((((-.((((-....)))).)))).)))))....)))).)))))))))) ( -21.60) >DroSec_CAF1 35300 94 + 1 AACAAACACA--AACACAAUAAGUACCCAGCUCAAUUUUGUGUUCUUUUUGG-GGUAAAAACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGC ..........--..........((((((...((((....(((((.((((((.-((((((....))).)))..))))))...))))).)))))))))) ( -18.00) >DroSim_CAF1 33543 93 + 1 AACCAACACA--AACACAACAAGUACCCAGCUCAAUUUUGUGUUCUUUUUGG-GGUAA-AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGU ..((((.(..--(((((((..(((.....))).....)))))))..).))))-(((.(-(....)).)))............(((((.....))))) ( -16.10) >DroEre_CAF1 35242 94 + 1 AAACAACACA--AACACAACAAGUACCCAGCUAAAUUUUGUGUUCUUUUUGGGGGAAA-AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGC ..........--..........((((((...............((((((((((..((.-...))..).)))).)))))(((......))).)))))) ( -18.30) >DroYak_CAF1 33972 95 + 1 AAACAACACAAAAACACAACAAGUACCCAGCUCAAUUUUGUGUUCUUUUUGG-GGAAA-AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGC ......................((((((...((((....(((((((((((((-.((((-....)))).)))).)))))....)))).)))))))))) ( -20.40) >consensus AAACAACACA__AACACAACAAGUACCCAGCUCAAUUUUGUGUUCUUUUUGG_GGAAA_AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGC ......................((((((...((((....(((((((((((((.(((((....))))).)))).)))))....)))).)))))))))) (-16.96 = -17.24 + 0.28)



| Location | 5,386,206 – 5,386,297 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5386206 91 + 23771897 UGUUCUUUUUGG-GGAAA-AACUUUUCGCCAGCAAAGACAAAACACCUUGGGGGUGCCAAGUCAGGCGGGUUAAGCCGAAAAAUAACAAUAAA ((((.(((((.(-(((((-....))))(((.((...(((....((((.....))))....)))..)).)))....)).))))).))))..... ( -22.80) >DroSec_CAF1 35338 92 + 1 UGUUCUUUUUGG-GGUAAAAACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGCCAAGUCAGGCGGGUUAAGCCGAAAAAUAACAAUAAA ((((.(((((.(-(((....)).(((.(((.((...(((....((((.....))))....)))..)).))).))))).))))).))))..... ( -20.30) >DroSim_CAF1 33581 91 + 1 UGUUCUUUUUGG-GGUAA-AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGUCAAGUCAGGCGGGUUAAGCCGAAAAAUAACAAUAAA ((((.(((((.(-(....-..(((...(((.((...(((...(((((.....)))))...)))..)).))).))))).))))).))))..... ( -22.10) >DroEre_CAF1 35280 92 + 1 UGUUCUUUUUGGGGGAAA-AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGCCAAGUCAGGCGGGUUAAGCCCAAAAAUAACAAUAAA ((((.((((((((.((((-....))))(((.((...(((....((((.....))))....)))..)).)))....)))))))).))))..... ( -26.90) >DroYak_CAF1 34012 91 + 1 UGUUCUUUUUGG-GGAAA-AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGCCAGGUCAGGCGGGUUAAGCCGAAAAAUAACAAUAAA ((((.(((((.(-(((((-....))))(((.((...(((....((((.....))))....)))..)).)))....)).))))).))))..... ( -23.60) >consensus UGUUCUUUUUGG_GGAAA_AACUUUUCACCAGCAAAGACAAAACACCUUGAGGGUGCCAAGUCAGGCGGGUUAAGCCGAAAAAUAACAAUAAA ((((.(((((((.(((((....)))))(((.((...(((....((((.....))))....)))..)).)))....)))))))..))))..... (-18.88 = -19.32 + 0.44)

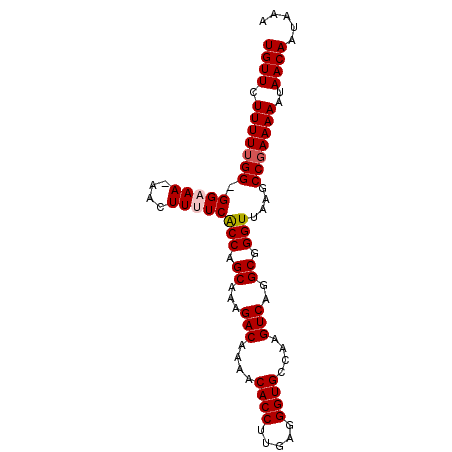

| Location | 5,386,206 – 5,386,297 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5386206 91 - 23771897 UUUAUUGUUAUUUUUCGGCUUAACCCGCCUGACUUGGCACCCCCAAGGUGUUUUGUCUUUGCUGGCGAAAAGUU-UUUCC-CCAAAAAGAACA .....((((.(((((.((...(((.((((.(((..((((((.....))))))..)))......))))....)))-....)-).))))).)))) ( -22.20) >DroSec_CAF1 35338 92 - 1 UUUAUUGUUAUUUUUCGGCUUAACCCGCCUGACUUGGCACCCUCAAGGUGUUUUGUCUUUGCUGGUGAAAAGUUUUUACC-CCAAAAAGAACA .....((((.(((((.((........((..(((..((((((.....))))))..)))...)).((((((.....))))))-))))))).)))) ( -22.30) >DroSim_CAF1 33581 91 - 1 UUUAUUGUUAUUUUUCGGCUUAACCCGCCUGACUUGACACCCUCAAGGUGUUUUGUCUUUGCUGGUGAAAAGUU-UUACC-CCAAAAAGAACA .....((((.(((((.((........((..(((..((((((.....))))))..)))...)).((((((....)-)))))-))))))).)))) ( -22.80) >DroEre_CAF1 35280 92 - 1 UUUAUUGUUAUUUUUGGGCUUAACCCGCCUGACUUGGCACCCUCAAGGUGUUUUGUCUUUGCUGGUGAAAAGUU-UUUCCCCCAAAAAGAACA .....((((.((((((((....(((.((..(((..((((((.....))))))..)))...)).)))((((....-)))).)))))))).)))) ( -29.00) >DroYak_CAF1 34012 91 - 1 UUUAUUGUUAUUUUUCGGCUUAACCCGCCUGACCUGGCACCCUCAAGGUGUUUUGUCUUUGCUGGUGAAAAGUU-UUUCC-CCAAAAAGAACA .....((((.(((((.((....(((.((..(((..((((((.....))))))..)))...)).)))((((....-)))))-).))))).)))) ( -21.60) >consensus UUUAUUGUUAUUUUUCGGCUUAACCCGCCUGACUUGGCACCCUCAAGGUGUUUUGUCUUUGCUGGUGAAAAGUU_UUUCC_CCAAAAAGAACA .....((((.(((((.(((((.(((.((..(((..((((((.....))))))..)))...)).)))...))).........))))))).)))) (-20.20 = -20.24 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:18 2006