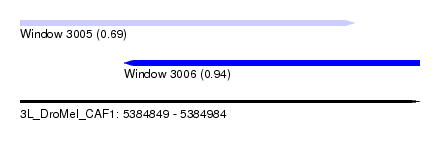
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,384,849 – 5,384,984 |
| Length | 135 |
| Max. P | 0.939465 |

| Location | 5,384,849 – 5,384,962 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
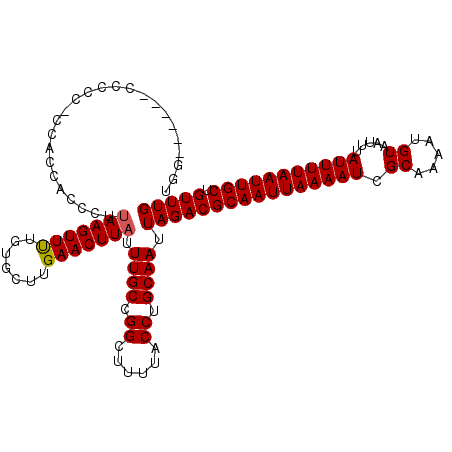
>3L_DroMel_CAF1 5384849 113 + 23771897 UGGUCG---CCCCCACCAUCACCCUUAAGUUUUGUGCUUGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG ((((..---.....)))).......(((((((.......))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).))))) ( -21.30) >DroSec_CAF1 33999 111 + 1 UGG-----CCCCCCACCGCCACCCUUAAGUUUUGUGCUUGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUUGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG (((-----(........))))....(((((((.......))))))).((((.((......)).)))).(((((((((((((((((((.....)))....))))))))))).))))) ( -25.30) >DroSim_CAF1 32236 116 + 1 UGGUCGCACCCCCCACCACCACCCUUAAGUUUUGUGCUUGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG ((((.(......).)))).......(((((((.......))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).))))) ( -21.20) >DroEre_CAF1 33970 98 + 1 ------------------CCACCCUUAAGUUUUGUGCUUGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG ------------------.......(((((((.......))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).))))) ( -19.40) >DroYak_CAF1 32667 112 + 1 UGUUUU---CCAUC-GCACCACCCUUAAGUUUUGUGCUUGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG ......---.....-((((.(..(....)..).))))..(((((((.((((.((......)).)))).)))..(((((((((((.((.....)).....))))))))))).)))). ( -19.40) >DroAna_CAF1 32093 99 + 1 -----------------UCCACCCUGAAGUUCUGUGCUUGAACUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG -----------------.........((((((.......))))))..((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).))))) ( -20.70) >consensus UGG______CCCCC_CCACCACCCUUAAGUUUUGUGCUUGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG .........................(((((((.......))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).))))) (-19.52 = -19.55 + 0.03)


| Location | 5,384,884 – 5,384,984 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
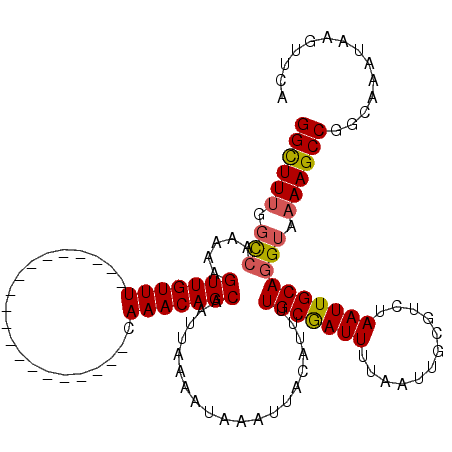
>3L_DroMel_CAF1 5384884 100 - 23771897 GGCUUUGGCCAAAAAGUUGUUU--------------------CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA ((((((.(((.....(((....--------------------..))).(((((((((((.((.......))...)))))))))))...........))).)))))).............. ( -24.30) >DroSec_CAF1 34032 100 - 1 GGCUUUGGCCAAAAAGUUGUUU--------------------CAAACAGCAAUUAAAAUAAAUUACAUUUUGCAAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA ((((((.(((.....(((....--------------------..))).(((((((....(((......)))(((((....)))))...))))))).))).)))))).............. ( -23.90) >DroEre_CAF1 33990 100 - 1 GGCUUUGGCCAAAAAGUUGUUU--------------------CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA ((((((.(((.....(((....--------------------..))).(((((((((((.((.......))...)))))))))))...........))).)))))).............. ( -24.30) >DroWil_CAF1 24880 117 - 1 GGUUU-GUUGUUAUUGUUGUUUUUUGUUUUUCGUUGUCUUCACAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGAAAA--ACUUUA (((((-(((((......(((..((((((((..(((((........)))))....))))))))..)))....))))).(((.(((((......))))).))))))))......--...... ( -21.40) >DroYak_CAF1 32701 100 - 1 GGCUUUGGCCAAAAAGUUGUUU--------------------CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA ((((((.(((.....(((....--------------------..))).(((((((((((.((.......))...)))))))))))...........))).)))))).............. ( -24.30) >DroAna_CAF1 32114 100 - 1 GGCUUUGGCCAAAAAGUUGUUU--------------------CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAGUUCA ((((((.(((.....(((....--------------------..))).(((((((((((.((.......))...)))))))))))...........))).)))))).............. ( -24.30) >consensus GGCUUUGGCCAAAAAGUUGUUU____________________CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA ((((((.(((.....(((((((.....................)))))))....................(((((((............)))))))))).)))))).............. (-20.33 = -20.42 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:10 2006