
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
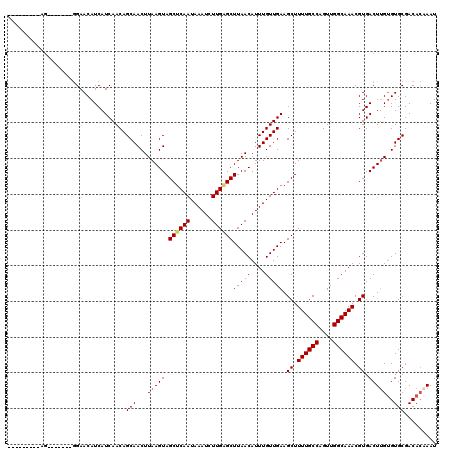
| Location | 5,379,203 – 5,379,378 |
| Length | 175 |
| Max. P | 0.913308 |

| Location | 5,379,203 – 5,379,307 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.68 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.87 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
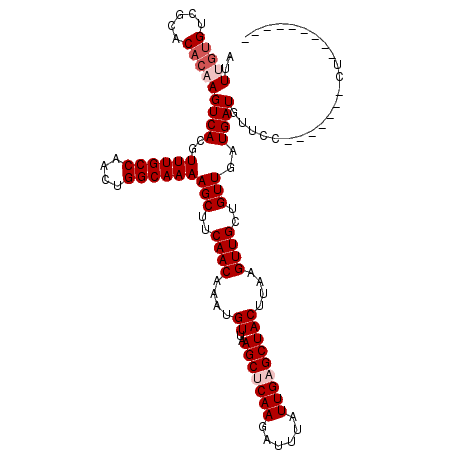
>3L_DroMel_CAF1 5379203 104 + 23771897 ---------AG-------GGAACAUCAUCAACAGCAACUUAAGUAGCUCAAUAAAUCUUGAGCUUAACAUUUGUUGAAGCUUUUGCCAGUUGGCAAACGUGACUUGUGUGCGACACAAAU ---------.(-------....)........((((((.......(((((((......)))))))......))))))..((.((((((....)))))).))...(((((.....))))).. ( -26.62) >DroSec_CAF1 28479 104 + 1 ---------AG-------GGAACAUCAUCAACAGCAACUUAAGUAGCACAAUAAAUCUUGAGCUUAACAUUUGUUGAAGCUUUUGCCAGUUGGCAAACGUGACUUGUGUGCGACACAAAU ---------.(-------....)..........(((...(((((..(((..........(((((((((....)))).)))))(((((....)))))..))))))))..)))......... ( -23.20) >DroSim_CAF1 26651 104 + 1 ---------AG-------GGAACAUCAUCAACAGCAACUUAAGUAGCUCAAUAAAUCUUGAGCUUAACAUUUGUUGAAGCUUUUGCCAGUUGGCAAACGUGACUUGUGUGCGACACAAAU ---------.(-------....)........((((((.......(((((((......)))))))......))))))..((.((((((....)))))).))...(((((.....))))).. ( -26.62) >DroEre_CAF1 28541 104 + 1 ---------AG-------GGAACAUCAUCAACAGCAACUUAAGUAGCUCAAUAAAUCUUGAGCUUAACAUUUGUUGAAGCUUUUGCCAGUUGGCAAACGUGACUUGUGUGCGACACAAAU ---------.(-------....)........((((((.......(((((((......)))))))......))))))..((.((((((....)))))).))...(((((.....))))).. ( -26.62) >DroYak_CAF1 27032 104 + 1 ---------AA-------GGAACAUCAUCAACAGCAACUUAAGUAGCUCAAUAAAUCUUGAGCUUAACAUUUGUUGAAGCUUUUGCCAGUUGGCAAACGUGACUUGUGUGCGACACCAAU ---------((-------(..((........((((((.......(((((((......)))))))......)))))).....((((((....)))))).))..)))(((.....))).... ( -25.82) >DroAna_CAF1 26485 120 + 1 GCGAGUGGCAGGGCAAAAAGAACAUCAUCAACAGCAACUUAAGUAGCUCAAUAAAUCUUGGGCUUAACAUUUGUUGAAGCUUUUGCCAGUUGGCAAACGUGACUGGAGUGCGACAGAAAU .(((.(((((((((.....((......))..((((((.......(((((((......)))))))......))))))..)).))))))).)))..........(((........))).... ( -30.22) >consensus _________AG_______GGAACAUCAUCAACAGCAACUUAAGUAGCUCAAUAAAUCUUGAGCUUAACAUUUGUUGAAGCUUUUGCCAGUUGGCAAACGUGACUUGUGUGCGACACAAAU .................................(((...(((((.((((((......))))))(((((....))))).((.((((((....)))))).)).)))))..)))......... (-23.64 = -23.87 + 0.22)


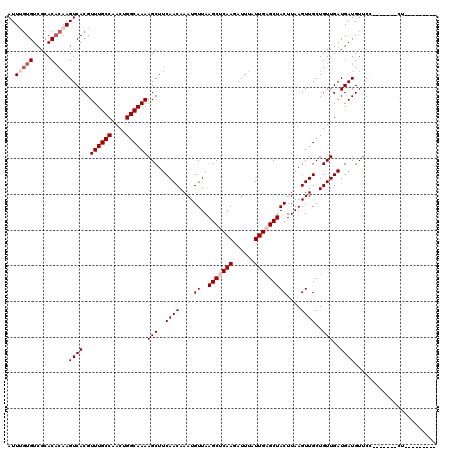
| Location | 5,379,203 – 5,379,307 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.68 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.28 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5379203 104 - 23771897 AUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAAGCUUCAACAAAUGUUAAGCUCAAGAUUUAUUGAGCUACUUAAGUUGCUGUUGAUGAUGUUCC-------CU--------- ..(((((.....)))))((((..((((((....))))))(((..((((....((..(((((((......)))))))))....))))..)))..)))).....-------..--------- ( -27.50) >DroSec_CAF1 28479 104 - 1 AUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAAGCUUCAACAAAUGUUAAGCUCAAGAUUUAUUGUGCUACUUAAGUUGCUGUUGAUGAUGUUCC-------CU--------- (((((.((.(((((.(((((...((((((....))))))(((((.(((....))))))))...)))))..))))).)).)))))..................-------..--------- ( -24.70) >DroSim_CAF1 26651 104 - 1 AUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAAGCUUCAACAAAUGUUAAGCUCAAGAUUUAUUGAGCUACUUAAGUUGCUGUUGAUGAUGUUCC-------CU--------- ..(((((.....)))))((((..((((((....))))))(((..((((....((..(((((((......)))))))))....))))..)))..)))).....-------..--------- ( -27.50) >DroEre_CAF1 28541 104 - 1 AUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAAGCUUCAACAAAUGUUAAGCUCAAGAUUUAUUGAGCUACUUAAGUUGCUGUUGAUGAUGUUCC-------CU--------- ..(((((.....)))))((((..((((((....))))))(((..((((....((..(((((((......)))))))))....))))..)))..)))).....-------..--------- ( -27.50) >DroYak_CAF1 27032 104 - 1 AUUGGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAAGCUUCAACAAAUGUUAAGCUCAAGAUUUAUUGAGCUACUUAAGUUGCUGUUGAUGAUGUUCC-------UU--------- ....(((.....)))..((((..((((((....))))))(((..((((....((..(((((((......)))))))))....))))..)))..)))).....-------..--------- ( -25.70) >DroAna_CAF1 26485 120 - 1 AUUUCUGUCGCACUCCAGUCACGUUUGCCAACUGGCAAAAGCUUCAACAAAUGUUAAGCCCAAGAUUUAUUGAGCUACUUAAGUUGCUGUUGAUGAUGUUCUUUUUGCCCUGCCACUCGC ......((.(((...((((.((.((((((....))))))((((.(((.((((.((......)).)))).)))))))......)).))))..((......)).........))).)).... ( -22.40) >consensus AUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAAGCUUCAACAAAUGUUAAGCUCAAGAUUUAUUGAGCUACUUAAGUUGCUGUUGAUGAUGUUCC_______CU_________ ..(((((.....)))))((((..((((((....))))))(((..((((....((..(((((((......)))))))))....))))..)))..))))....................... (-23.28 = -24.28 + 1.00)

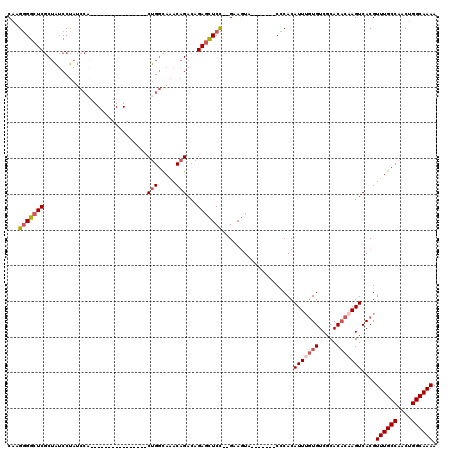
| Location | 5,379,267 – 5,379,378 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.93 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5379267 111 - 23771897 CAUGGGGCUCGCUAUCCUAUCCAACCAAAUGCGAGGGGACUGGCAAACAGACAGAGCUCC--GAAGUA-------CCCACAUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAA ..((((((((....((((.((((......)).)).))))(((.....)))...)))))))--).....-------.....(((((((.....)))))))....((((((....)))))). ( -34.80) >DroSec_CAF1 28543 95 - 1 CAAGGGGCUCGCUAUCCUAUCCA----------------CUGGCAAACAGACAGAGCUCC--GAAGUA-------CCCACAUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAA ...(((((((.((..((......----------------..)).....))...)))))))--......-------.....(((((((.....)))))))....((((((....)))))). ( -26.40) >DroSim_CAF1 26715 95 - 1 CAAGGGGCUCGCUAUCCUAUCCA----------------CUGGCAAACAGACAGAGCUCC--GAAGUA-------CCCACAUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAA ...(((((((.((..((......----------------..)).....))...)))))))--......-------.....(((((((.....)))))))....((((((....)))))). ( -26.40) >DroEre_CAF1 28605 107 - 1 CAAGGGGCUCGCUAUCCUAUCCAACCAAAUGCGA----GCUGGCAAACAGACAGAGCUCC--GAAGUA-------CCCACAUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAA ...(((((((((..................))..----((((.....))).).)))))))--......-------.....(((((((.....)))))))....((((((....)))))). ( -29.57) >DroYak_CAF1 27096 107 - 1 CAAGGGGCUCGCUAUCCUAUCCCACCAAAUGCGA----ACUGGCAAACAGACAGAGCUCC--GAAGUA-------CUCACAUUGGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAA ...(((((((((..................))..----.(((.....)))...)))))))--......-------.........(((.(........).))).((((((....)))))). ( -27.47) >DroAna_CAF1 26565 99 - 1 C--GUGGCUCACAUGCCGGU-------------------CGGUUAGACAGACAGACUUCUUGGUAAUAUCCAGCUCACACAUUUCUGUCGCACUCCAGUCACGUUUGCCAACUGGCAAAA (--((((((....(((..((-------------------(.....))).((((((.....(((......)))(......)...)))))))))....)))))))((((((....)))))). ( -30.20) >consensus CAAGGGGCUCGCUAUCCUAUCCA________________CUGGCAAACAGACAGAGCUCC__GAAGUA_______CCCACAUUUGUGUCGCACACAAGUCACGUUUGCCAACUGGCAAAA ...((((((((.....)......................(((.....)))...)))))))....................(((((((.....)))))))....((((((....)))))). (-17.88 = -18.93 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:05 2006