
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
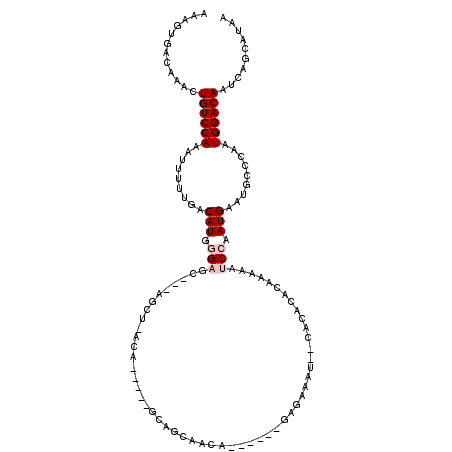
| Location | 5,366,020 – 5,366,123 |
| Length | 103 |
| Max. P | 0.658072 |

| Location | 5,366,020 – 5,366,123 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -6.12 |
| Energy contribution | -6.78 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5366020 103 + 23771897 AAAGUGACAAACUGUCCAAAUUUUUGACAUGGGAGC---AGCU-CCA-----GCAGCAACA------GAGAAAU--CACACACAAAAAUCCAAUGAAUGCCCAAUGGACAAUCAGCAUAA ...(((......((((((.........(((.(((((---.((.-...-----)).))....------((....)--)...........))).))).........)))))).....))).. ( -18.87) >DroVir_CAF1 31424 102 + 1 AAAGUGACAAACUGUCCACAUUUUUGACAUGGGCAU---ACACCAAA----AGUAAAAAAAU--------AAAA---AGGAAAACAAUUCAAAUGAAUGCCCAAUGGACAAUCACUAACA ..(((((.....((((((((....))...(((((.(---((......----.))).......--------....---.........((((....))))))))).)))))).))))).... ( -21.50) >DroSim_CAF1 13371 105 + 1 AAAGUGACAAACUGUCCAAAUUUUUGACAUGGGAGC---AGCU-CUA---GAGCAGCAACA------GAGAAAU--CACACACAAAAAUCCAAUGAAUGCCCAAUGGACAAUCAGCAUAA ...(((......((((((.........(((.(((((---.(((-...---.))).))....------((....)--)...........))).))).........)))))).....))).. ( -20.47) >DroYak_CAF1 13742 106 + 1 AAAGUGACAAACUGUCCAAAUUUUUGACAUGGGAGCAGCAGCU-UCA-----GUAGCAACA------GAGAAAU--CACACACAAAAAUCCAAUGAAUGCCCAAUGGACAAUCAGCAUAA ...(((......((((((.(((((((.....(((((....)))-)).-----.........------((....)--).....)))))))....((......)).)))))).....))).. ( -19.60) >DroMoj_CAF1 29951 110 + 1 AAAGUGACAAACUGUCCAAAUUUUUGACAUG-GCAU---AUAGCAAA-----GCAACAAAAGCAAUUGC-AAAAAACCCGAAAUGAAUUCAAAUGAAUGCCCAAUGGACAGUCACUAACA ..((((((....((((((.....(((....(-((((---...((((.-----((.......))..))))-.........(((.....)))......)))))))))))))))))))).... ( -23.80) >DroAna_CAF1 13541 103 + 1 AAAGUGACAAACUGUCCAAAAUUUUGACAUGGGAGC---AGCC-ACAUCAGAGUAGCUG-A------AAGAAAU--CA----CAAAAAUCCAAUGAAUGCCCAAUGGACAAUCAACAUAA ...((((....((.((....(((((((..(((....---..))-)..)))))))....)-)------.))...)--))----).....((((.((......)).))))............ ( -17.10) >consensus AAAGUGACAAACUGUCCAAAUUUUUGACAUGGGAGC___AGCU_ACA_____GCAGCAACA______GAGAAAU__CACACACAAAAAUCCAAUGAAUGCCCAAUGGACAAUCAGCAUAA ............((((((.........(((.(((......................................................))).))).........)))))).......... ( -6.12 = -6.78 + 0.67)

| Location | 5,366,020 – 5,366,123 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5366020 103 - 23771897 UUAUGCUGAUUGUCCAUUGGGCAUUCAUUGGAUUUUUGUGUGUG--AUUUCUC------UGUUGCUGC-----UGG-AGCU---GCUCCCAUGUCAAAAAUUUGGACAGUUUGUCACUUU ..((((.((..(((((.(((....))).)))))..))))))(((--((....(------(((.((.((-----...-.)).---))..(((...........)))))))...)))))... ( -22.20) >DroVir_CAF1 31424 102 - 1 UGUUAGUGAUUGUCCAUUGGGCAUUCAUUUGAAUUGUUUUCCU---UUUU--------AUUUUUUUACU----UUUGGUGU---AUGCCCAUGUCAAAAAUGUGGACAGUUUGUCACUUU ....((((((((((((((((((((.(((..(((.((.......---....--------.......)).)----))..))).---)))))))..........)))))))....)))))).. ( -25.27) >DroSim_CAF1 13371 105 - 1 UUAUGCUGAUUGUCCAUUGGGCAUUCAUUGGAUUUUUGUGUGUG--AUUUCUC------UGUUGCUGCUC---UAG-AGCU---GCUCCCAUGUCAAAAAUUUGGACAGUUUGUCACUUU ..((((.((..(((((.(((....))).)))))..))))))(((--((....(------(((.((.(((.---...-))).---))..(((...........)))))))...)))))... ( -23.80) >DroYak_CAF1 13742 106 - 1 UUAUGCUGAUUGUCCAUUGGGCAUUCAUUGGAUUUUUGUGUGUG--AUUUCUC------UGUUGCUAC-----UGA-AGCUGCUGCUCCCAUGUCAAAAAUUUGGACAGUUUGUCACUUU ....((.(((((((((.........(((.(((.(((.(((.(..--((.....------.))..))))-----.))-)((....))))).))).........))))))))).))...... ( -22.57) >DroMoj_CAF1 29951 110 - 1 UGUUAGUGACUGUCCAUUGGGCAUUCAUUUGAAUUCAUUUCGGGUUUUUU-GCAAUUGCUUUUGUUGC-----UUUGCUAU---AUGC-CAUGUCAAAAAUUUGGACAGUUUGUCACUUU ....((((((((((((...(((((.....((((.....))))(((.....-((((..((.......))-----.))))...---..))-)))))).......))))))....)))))).. ( -28.20) >DroAna_CAF1 13541 103 - 1 UUAUGUUGAUUGUCCAUUGGGCAUUCAUUGGAUUUUUG----UG--AUUUCUU------U-CAGCUACUCUGAUGU-GGCU---GCUCCCAUGUCAAAAUUUUGGACAGUUUGUCACUUU ...((..(((((((((...(((((((((.........)----))--)......------.-(((((((......))-))))---).....))))).......)))))))))...)).... ( -25.90) >consensus UUAUGCUGAUUGUCCAUUGGGCAUUCAUUGGAUUUUUGUGUGUG__AUUUCUC______UGUUGCUAC_____UGU_AGCU___GCUCCCAUGUCAAAAAUUUGGACAGUUUGUCACUUU .......(((((((((...(((((((....)).............................................(((....)))...))))).......)))))))))......... (-11.48 = -11.87 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:58 2006