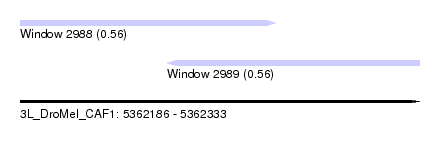
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,362,186 – 5,362,333 |
| Length | 147 |
| Max. P | 0.558222 |

| Location | 5,362,186 – 5,362,280 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -11.90 |
| Consensus MFE | -6.90 |
| Energy contribution | -7.54 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5362186 94 + 23771897 AAAACCAAACAACGAUACGAACUGCAACACUAAGCAAGGAUAUACAAAAUUAAAGUUCCAUCAACGGAACUACAUACAUAUGUAGUCCAUAGAA .............(((..(((((((........))..(......)........))))).)))...(((.((((((....)))))))))...... ( -15.70) >DroSec_CAF1 11376 85 + 1 AAAACCAAACAA-----CAAGCUGCAAUACUAAGCAAGGACAUACAAAAUUAAAGUUCCAUCAACGAAACUAC----AUAUGUAGUCUAUAGAA ............-----...(((.........)))..((((.((((.......((((.(......).))))..----...))))))))...... ( -8.60) >DroSim_CAF1 9462 90 + 1 AAAACCAAACAACGGAACGAGCUGCAACACUAAGCAAGGACAUACAAAAUUAAAGUUCCACCAACUGAAGUAC----AUAUGUAGUCCAUAGAA ....((.......)).....(((.........)))..((((.((((..(((..((((.....))))..)))..----...))))))))...... ( -11.50) >DroEre_CAF1 10043 86 + 1 AAAACCAACCAACGCAACGAGCUGCAACACUCAGCCAAGACAUACAAAAUUGAAGUUCCAUCAACGGAACUA--------UGUAGUAAAUAGUA ....................((((((..............((........)).((((((......)))))).--------))))))........ ( -13.40) >DroYak_CAF1 9848 79 + 1 AAAACCAACCAAAGGAACGAGCUGCAACACUCUGUGAAGACAUACAAAAUUGAAGUUCCAUACACGAAACUA--------UGUAG-------AA .............((((((((........)))((((....))))..........))))).((((........--------)))).-------.. ( -10.30) >consensus AAAACCAAACAACGGAACGAGCUGCAACACUAAGCAAGGACAUACAAAAUUAAAGUUCCAUCAACGGAACUAC____AUAUGUAGUCCAUAGAA ....................((((((...((......))..............((((((......)))))).........))))))........ ( -6.90 = -7.54 + 0.64)



| Location | 5,362,240 – 5,362,333 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -17.62 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5362240 93 - 23771897 --------------------UCCUUAUCUGCAUUAACUUAAAAGCAGAAAGUUUUCUCAAAAAUGGUAGCUCUUUCUAUGGACUACAUAUGUAUGUAGUUCCGUUGAUGGAAC --------------------..........(((((((........((((((..((..((....))..))..))))))..(((((((((....))))))))).))))))).... ( -21.90) >DroSec_CAF1 11425 107 - 1 AA-AAUGGGAGUUCAUGUAUUUCGUAUCUGCAUUAACUUAAAAGCAGAAAGUUUCCUAA-AAAUGGUAGCUCUUUCUAUAGACUACAUAU----GUAGUUUCGUUGAUGGAAC ..-.......(((((((.....))).....(((((((........((((((...((...-....)).....))))))..(((((((....----))))))).))))))))))) ( -21.10) >DroSim_CAF1 9516 109 - 1 AAAAAUGGGAGUUCAUGUAUUUCUUAUCUGCAUUAACUUAAAAGCAGAAAGUUUUCUAAAAAAUGGUAGCUCUUUCUAUGGACUACAUAU----GUACUUCAGUUGGUGGAAC .......(((((.(((((....(((.(((((.((......)).))))))))..............((((.(((......)))))))))))----).)))))............ ( -20.50) >consensus AA_AAUGGGAGUUCAUGUAUUUCUUAUCUGCAUUAACUUAAAAGCAGAAAGUUUUCUAAAAAAUGGUAGCUCUUUCUAUGGACUACAUAU____GUAGUUCCGUUGAUGGAAC ..............................(((((((........((((((...((........)).....))))))..(((((((........))))))).))))))).... (-17.62 = -17.07 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:54 2006