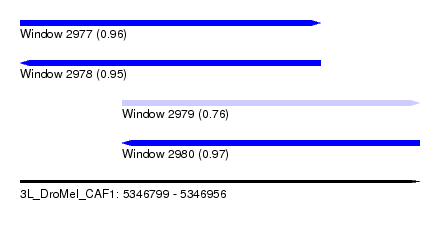
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,346,799 – 5,346,956 |
| Length | 157 |
| Max. P | 0.965500 |

| Location | 5,346,799 – 5,346,917 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.80 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346799 118 + 23771897 UUACCAUGGGGGCGUAUACGUGUUAAGCGUAGGCAAAAGAUAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGACAGUUUUUAC--CCCAGUCCAUAGACCACAGCAACAUCUC ......(((((.....(((((.....)))))(((((..((((((((((((((...)))))))))))).))..)))))............)--))))(((....))).............. ( -36.40) >DroSec_CAF1 94720 111 + 1 UUACCAUGGGGGCGUAUACGUGUUAAGCGUAGGCAAAAGUUAAAUAUUGUCCUUUGGACAAUAUUGGCUCAGUUGCCCGGCUGUUUUUCC--CCCAGUCC-------ACAGCAACAUCUC ......((((((....(((((.....)))))(((((.((((((.((((((((...))))))))))))))...)))))...........))--))))....-------............. ( -35.60) >DroSim_CAF1 96483 111 + 1 UUACCAUGGGGGCGUAUACGUGUUAAGCGUAGGCAAAAGAUAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGGCUGUUUUUCC--CUCAGUCC-------ACAGCAACAUCUC ......((((((....(((((.....)))))(((((..((((((((((((((...)))))))))))).))..)))))...........))--))))....-------............. ( -34.40) >DroEre_CAF1 94808 120 + 1 UUACCAUGGGGGCGUAUACGUGUUAAGCGUAGGCAAGAGAUAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGGCUUUUUUGCCAGCUCUGUCCGACAUCUACAGCUACAUCUC ......(((((((((.(((((.....))))).))..(((.((((((((((((...)))))))))))))))....))))(((......)))........)))................... ( -39.20) >DroYak_CAF1 103520 90 + 1 UUACCAUGGGGGCGUAUACGUGUUAAGCGUAGGCAAAAGAUAAAUAUUGUCCUUUGGACAAUAUUUACCCAGUUGCCCGGCUUUUUU------------------------------CCC .......((((((...(((((.....)))))(((((..(.((((((((((((...)))))))))))).)...)))))..))).....------------------------------))) ( -28.40) >DroAna_CAF1 96909 93 + 1 UUACCAUGGGGGCGUAUACGUGUUAAGCGUAGGCAGAAGACAAAUAUUGUCCCUUCAACAAUAUUUGCUCGGCU------CUGUUUUCCC-----AC----------------UCAUCUC ......((((((....(((((.....)))))((((((.((((((((((((.......)))))))))).))...)------))))))))))-----).----------------....... ( -25.70) >consensus UUACCAUGGGGGCGUAUACGUGUUAAGCGUAGGCAAAAGAUAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGGCUGUUUUUCC__C_CAGUCC_______ACAGCAACAUCUC .........((((((.(((((.....))))).))....(((((((((((((.....))))))))))).))....)))).......................................... (-22.74 = -23.80 + 1.06)

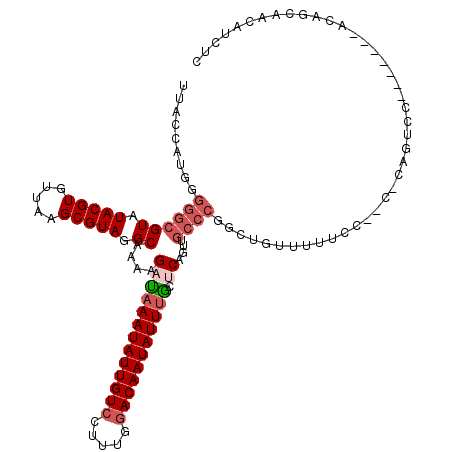
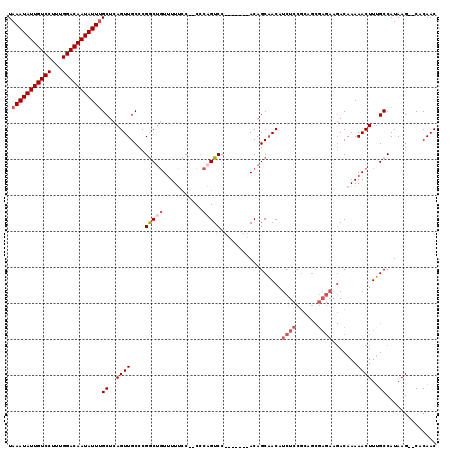
| Location | 5,346,799 – 5,346,917 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -18.77 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346799 118 - 23771897 GAGAUGUUGCUGUGGUCUAUGGACUGGG--GUAAAAACUGUCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUAUCUUUUGCCUACGCUUAACACGUAUACGCCCCCAUGGUAA ......(((((((((((....))).(((--((..........((((((..((..(((((((((((...))))))))))).))..))))))(((.......)))....))))))))))))) ( -39.90) >DroSec_CAF1 94720 111 - 1 GAGAUGUUGCUGU-------GGACUGGG--GGAAAAACAGCCGGGCAACUGAGCCAAUAUUGUCCAAAGGACAAUAUUUAACUUUUGCCUACGCUUAACACGUAUACGCCCCCAUGGUAA ......(((((((-------((...(((--.(..........((((((..(((..((((((((((...))))))))))...)))))))))(((.......)))...).)))))))))))) ( -33.50) >DroSim_CAF1 96483 111 - 1 GAGAUGUUGCUGU-------GGACUGAG--GGAAAAACAGCCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUAUCUUUUGCCUACGCUUAACACGUAUACGCCCCCAUGGUAA ......(((((((-------((.....(--((......(((.((((((..((..(((((((((((...))))))))))).))..))))))..))).....((....)))))))))))))) ( -35.70) >DroEre_CAF1 94808 120 - 1 GAGAUGUAGCUGUAGAUGUCGGACAGAGCUGGCAAAAAAGCCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUAUCUCUUGCCUACGCUUAACACGUAUACGCCCCCAUGGUAA ...(((..(.((((.((((.(....((((.(((......)))((((((..((..(((((((((((...))))))))))).))..))))))..))))..))))).)))).)..)))..... ( -37.00) >DroYak_CAF1 103520 90 - 1 GGG------------------------------AAAAAAGCCGGGCAACUGGGUAAAUAUUGUCCAAAGGACAAUAUUUAUCUUUUGCCUACGCUUAACACGUAUACGCCCCCAUGGUAA (((------------------------------....((((.((((((..(((((((((((((((...))))))))))))))).))))))..))))....((....))..)))....... ( -34.40) >DroAna_CAF1 96909 93 - 1 GAGAUGA----------------GU-----GGGAAAACAG------AGCCGAGCAAAUAUUGUUGAAGGGACAAUAUUUGUCUUCUGCCUACGCUUAACACGUAUACGCCCCCAUGGUAA .......----------------((-----(((....(((------((....((((((((((((.....)))))))))))).)))))..((((.......))))......)))))..... ( -23.20) >consensus GAGAUGUUGCUGU_______GGACUG_G__GGAAAAACAGCCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUAUCUUUUGCCUACGCUUAACACGUAUACGCCCCCAUGGUAA ...........................................(((((..((...(((((((((.....)))))))))..))..)))))((((.......))))...(((.....))).. (-18.77 = -18.92 + 0.14)

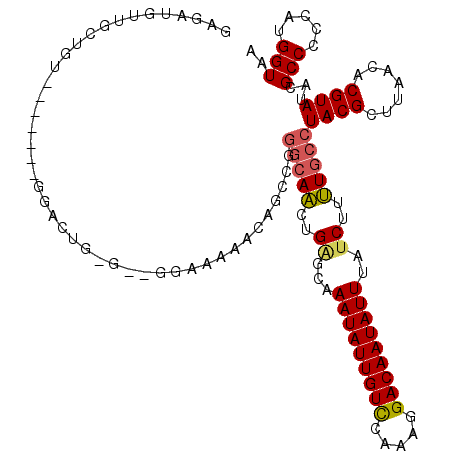
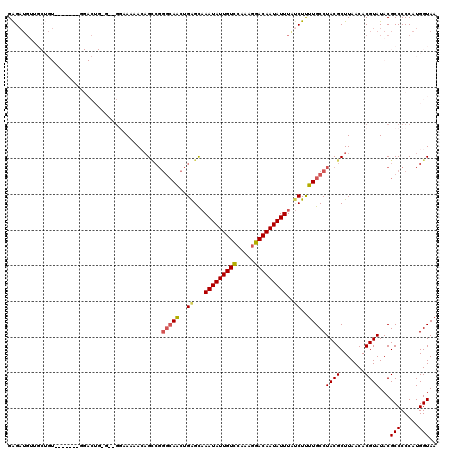
| Location | 5,346,839 – 5,346,956 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -15.36 |
| Energy contribution | -17.18 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346839 117 + 23771897 UAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGACAGUUUUUAC--CCCAGUCCAUAGACCACAGCAACAUCUCCGCAGCGAGAAGACAAAAACUUUGCCAUAAGCGCACAAC .(((((((((((...)))))))))))(((..(((((......((....))--....(((....)))....))))).((((......))))...................)))....... ( -22.70) >DroSec_CAF1 94760 108 + 1 UAAAUAUUGUCCUUUGGACAAUAUUGGCUCAGUUGCCCGGCUGUUUUUCC--CCCAGUCC-------ACAGCAACAUCUCCGCAGCGAGAAGACAAAAACUUUGCCAUAAG--CACAAC ..((((((((((...))))))))))(((...(((((..(((((.......--..))))).-------...))))).((((......)))).............))).....--...... ( -28.40) >DroSim_CAF1 96523 98 + 1 UAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGGCUGUUUUUCC--CUCAGUCC-------ACAGCAACAUCUCCG----------ACAAAAACUUUGCCAUAAG--CACAAC .(((((((((((...)))))))))))(((..(((((..(((((.......--..))))).-------...)))))......(----------.(((.....))).)...))--)..... ( -23.50) >DroEre_CAF1 94848 117 + 1 UAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGGCUUUUUUGCCAGCUCUGUCCGACAUCUACAGCUACAUCUCCAAUGCGAGAAGACAAAAACUUGGCCAUAAG--CACAAC .(((((((((((...)))))))))))(((..((.(((.(((......))).....((((.(....)..........((((......)))).)))).......))).)).))--)..... ( -27.50) >consensus UAAAUAUUGUCCUUUGGACAAUAUUUGCUCAGUUGCCCGGCUGUUUUUCC__CCCAGUCC_______ACAGCAACAUCUCCGCAGCGAGAAGACAAAAACUUUGCCAUAAG__CACAAC .(((((((((((...)))))))))))((..((((....(((((...........))))).................((((......)))).......))))..)).............. (-15.36 = -17.18 + 1.81)

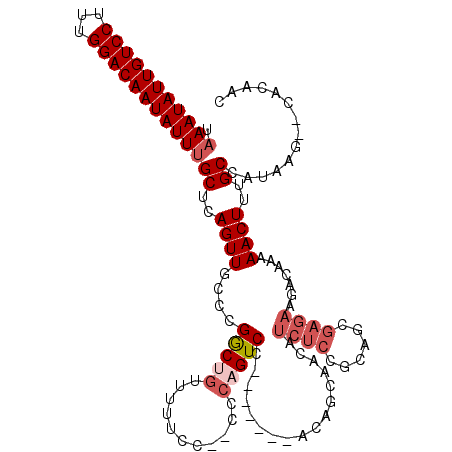
| Location | 5,346,839 – 5,346,956 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -25.99 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346839 117 - 23771897 GUUGUGCGCUUAUGGCAAAGUUUUUGUCUUCUCGCUGCGGAGAUGUUGCUGUGGUCUAUGGACUGGG--GUAAAAACUGUCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUA .....((((.....))..(((((((((((((.......))))))...(((.(((((....))))).)--))))))))).((((....))))))(((((((((((...))))))))))). ( -37.10) >DroSec_CAF1 94760 108 - 1 GUUGUG--CUUAUGGCAAAGUUUUUGUCUUCUCGCUGCGGAGAUGUUGCUGU-------GGACUGGG--GGAAAAACAGCCGGGCAACUGAGCCAAUAUUGUCCAAAGGACAAUAUUUA ......--....((((...((((((.(((((...(..(((........))).-------.)...)))--))))))))...(((....))).))))(((((((((...)))))))))... ( -33.70) >DroSim_CAF1 96523 98 - 1 GUUGUG--CUUAUGGCAAAGUUUUUGU----------CGGAGAUGUUGCUGU-------GGACUGAG--GGAAAAACAGCCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUA .....(--((((((((((.....))))----------)).....((((((.(-------((.(((..--.......)))))))))))))))))(((((((((((...))))))))))). ( -31.60) >DroEre_CAF1 94848 117 - 1 GUUGUG--CUUAUGGCCAAGUUUUUGUCUUCUCGCAUUGGAGAUGUAGCUGUAGAUGUCGGACAGAGCUGGCAAAAAAGCCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUA .....(--((((..(((..((((((((((((.......))))))...........((((((......)))))).))))))..)))...)))))(((((((((((...))))))))))). ( -38.10) >consensus GUUGUG__CUUAUGGCAAAGUUUUUGUCUUCUCGCUGCGGAGAUGUUGCUGU_______GGACUGAG__GGAAAAACAGCCGGGCAACUGAGCAAAUAUUGUCCAAAGGACAAUAUUUA (((........(((((((.(((((((...........))))))).)))))))........)))...............(((((....))).))(((((((((((...))))))))))). (-25.99 = -26.12 + 0.13)

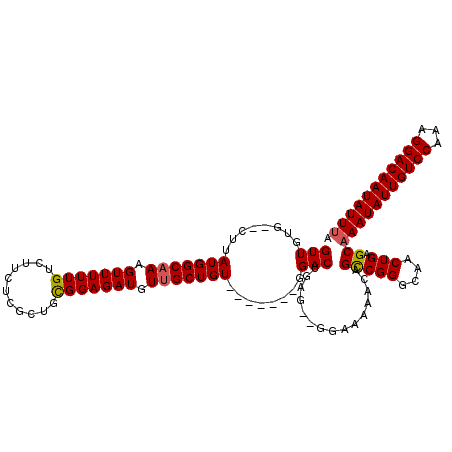
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:46 2006