
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,346,227 – 5,346,397 |
| Length | 170 |
| Max. P | 0.999216 |

| Location | 5,346,227 – 5,346,339 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -22.96 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346227 112 + 23771897 UUUGGGCAAUUUUGAGAGACUGCGCCAAAGGUCGGCGCGUCUAAAUCAAUGCGCAAAAAAUGCAGCCAU---CGAGUGGAGUUCCCGAAAAAUGGGCAAAAGCUGUAGUAAU---CCC ((((.(((...((((.((((.(((((.......)))))))))...))))))).))))...((((((..(---(.....))...((((.....)))).....)))))).....---... ( -33.90) >DroSec_CAF1 94132 94 + 1 UUUGGGCAAUUUUGAGAGACUGCGCCAAAGGUCGGCGCGUCUAAAUCAAUGCGCAAAAAAUGCAGCCAU---CGAGUGGAGCUCC------------------AGUAGUAAU---CCC ....(((....((((.((((.(((((.......)))))))))...))))...(((.....))).)))..---..(.(((....))------------------).)......---... ( -28.00) >DroSim_CAF1 95904 94 + 1 UUUGGGCAAUUUUGAGAGACUGCGCCAAAGGUCGGCGCGUCUAAAUCAAUGCGCAAAAAAUGCAGCCAU---CGAGUGGAGUUCC------------------AGUAGUAAU---CCC ....(((....((((.((((.(((((.......)))))))))...))))...(((.....))).)))..---..(.(((....))------------------).)......---... ( -28.00) >DroEre_CAF1 94158 118 + 1 UUUGGGCAAUUUUGAAAGACUGCGCCAAAGGUCGGCGCGUCUAAAUCAAUGCGGAAAAAAUGCAGCCAUCCUCCAGUGGAGCUCCAGAAAAAUGGGCAGAAGCUGUAGUAAUCGGCGG ...........((((.((((.(((((.......)))))))))...))))(((.((.....((((((..(((......)))((.(((......)))))....))))))....)).))). ( -37.30) >DroYak_CAF1 102846 99 + 1 UUUGGGCAAUUUUGAGAGACUGCGCCAAAGGUCGGUGCGUCUAAAUCAAUGCGGAAAAAAUGCAGC------------------CAGAAAAAUGGGCAAAAGCUCUAGUA-UAAGCCC ....(((....((((.((((.(((((.......)))))))))...))))((((.......))))))------------------)........((((....((....)).-...)))) ( -29.10) >consensus UUUGGGCAAUUUUGAGAGACUGCGCCAAAGGUCGGCGCGUCUAAAUCAAUGCGCAAAAAAUGCAGCCAU___CGAGUGGAGCUCC_GAAAAAUGGGCA_AAGCUGUAGUAAU___CCC ....(((....((((.((((.(((((.......)))))))))...))))((((.......)))))))................................................... (-22.96 = -23.00 + 0.04)

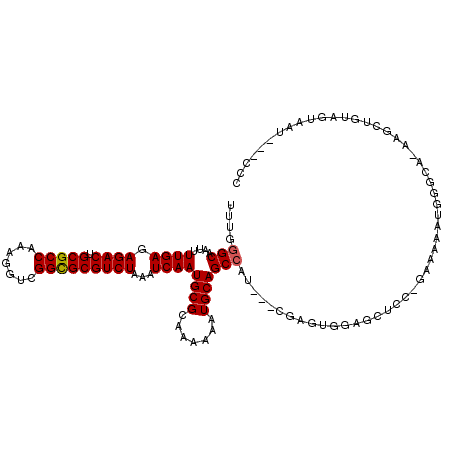
| Location | 5,346,227 – 5,346,339 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -19.12 |
| Energy contribution | -20.12 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346227 112 - 23771897 GGG---AUUACUACAGCUUUUGCCCAUUUUUCGGGAACUCCACUCG---AUGGCUGCAUUUUUUGCGCAUUGAUUUAGACGCGCCGACCUUUGGCGCAGUCUCUCAAAAUUGCCCAAA (((---.........((....(((......(((((.......))))---).))).(((.....))))).((((...(((((((((((...))))))).)))).)))).....)))... ( -32.90) >DroSec_CAF1 94132 94 - 1 GGG---AUUACUACU------------------GGAGCUCCACUCG---AUGGCUGCAUUUUUUGCGCAUUGAUUUAGACGCGCCGACCUUUGGCGCAGUCUCUCAAAAUUGCCCAAA (((---........(------------------((....))).(((---(((.(.(((.....))))))))))...(((((((((((...))))))).))))..........)))... ( -28.40) >DroSim_CAF1 95904 94 - 1 GGG---AUUACUACU------------------GGAACUCCACUCG---AUGGCUGCAUUUUUUGCGCAUUGAUUUAGACGCGCCGACCUUUGGCGCAGUCUCUCAAAAUUGCCCAAA (((---........(------------------((....))).(((---(((.(.(((.....))))))))))...(((((((((((...))))))).))))..........)))... ( -28.40) >DroEre_CAF1 94158 118 - 1 CCGCCGAUUACUACAGCUUCUGCCCAUUUUUCUGGAGCUCCACUGGAGGAUGGCUGCAUUUUUUCCGCAUUGAUUUAGACGCGCCGACCUUUGGCGCAGUCUUUCAAAAUUGCCCAAA ..((.((......(((((((((((((......))).))(((...)))))).))))).......)).)).((((...(((((((((((...))))))).)))).))))........... ( -33.42) >DroYak_CAF1 102846 99 - 1 GGGCUUA-UACUAGAGCUUUUGCCCAUUUUUCUG------------------GCUGCAUUUUUUCCGCAUUGAUUUAGACGCACCGACCUUUGGCGCAGUCUCUCAAAAUUGCCCAAA ((((...-....((((...(((((((....((.(------------------(.(((.(((..((......))...))).)))))))....))).)))).)))).......))))... ( -25.24) >consensus GGG___AUUACUACAGCUU_UGCCCAUUUUUC_GGAACUCCACUCG___AUGGCUGCAUUUUUUGCGCAUUGAUUUAGACGCGCCGACCUUUGGCGCAGUCUCUCAAAAUUGCCCAAA (((.................................................((.(((.....))))).((((...(((((((((((...))))))).)))).)))).....)))... (-19.12 = -20.12 + 1.00)



| Location | 5,346,302 – 5,346,397 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -10.42 |
| Energy contribution | -12.42 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346302 95 + 23771897 GAGUUCCCGAAAAAUGGGCAAAAGCUGUAGUAAU---CCCAACUGCAGCUCAAUCAAAAUGCUCAGCGGAAUUUAAAUCAAAAUUCAAAUUGAGCAUC ((...((((.....))))....(((((((((...---....)))))))))...))...((((((((..((((((......))))))...)))))))). ( -27.90) >DroSec_CAF1 94207 76 + 1 GAGCUCC------------------AGUAGUAAU---CCCACCUGCAGUUCAACCAAAAUGCUCAGAGGAAUUUAAAUCAAAA-UCAAAUUGAGCAUC (((((.(------------------((..(....---..)..))).))))).......((((((((..((.(((.....))).-))...)))))))). ( -16.30) >DroSim_CAF1 95979 76 + 1 GAGUUCC------------------AGUAGUAAU---CCCACCUGCAGCUCAACCAAAAUGCUCAGCGGAAUUUAAAUCAAAA-UCAAAUUGAGCAUC (((((.(------------------((..(....---..)..))).))))).......((((((((..((.(((.....))).-))...)))))))). ( -16.30) >DroEre_CAF1 94236 97 + 1 GAGCUCCAGAAAAAUGGGCAGAAGCUGUAGUAAUCGGCGGGAUUACAGCUCUACCCAAAUGCUCAUCGGAAUUUAAAGCAAAA-UCGAAUUGAGCAUC ..............((((.(((.((((((((..........))))))))))).)))).((((((((((..((((......)))-))))..))))))). ( -27.00) >DroYak_CAF1 102912 90 + 1 ------CAGAAAAAUGGGCAAAAGCUCUAGUA-UAAGCCCUACUACAGCUCUACCAAAAUGCUCAUCGGAAUUUGAAUCGAAA-UCAAAUUGAACAUC ------.......(((((((..((((.(((((-.......))))).)))).........)))))))...(((((((.......-)))))))....... ( -15.70) >consensus GAGCUCC_GAAAAAUGGGCA_AAGCUGUAGUAAU___CCCAACUGCAGCUCAACCAAAAUGCUCAGCGGAAUUUAAAUCAAAA_UCAAAUUGAGCAUC .......................((((((((..........)))))))).........((((((((..((.(((......))).))...)))))))). (-10.42 = -12.42 + 2.00)

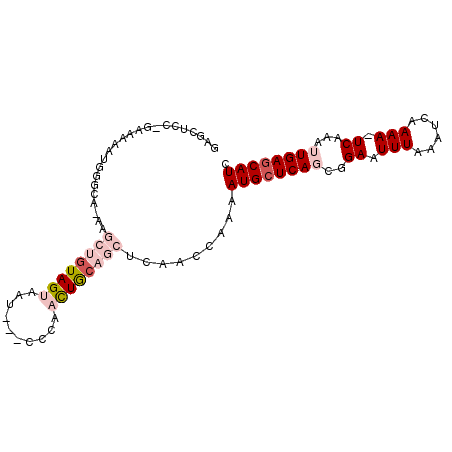

| Location | 5,346,302 – 5,346,397 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -8.46 |
| Energy contribution | -8.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5346302 95 - 23771897 GAUGCUCAAUUUGAAUUUUGAUUUAAAUUCCGCUGAGCAUUUUGAUUGAGCUGCAGUUGGG---AUUACUACAGCUUUUGCCCAUUUUUCGGGAACUC ((((((((....((((((......))))))...))))))))......((((((.(((....---...))).))))))...(((.......)))..... ( -24.70) >DroSec_CAF1 94207 76 - 1 GAUGCUCAAUUUGA-UUUUGAUUUAAAUUCCUCUGAGCAUUUUGGUUGAACUGCAGGUGGG---AUUACUACU------------------GGAGCUC ((((((((....((-(((......)))))....))))))))......((.((.(((.(((.---....)))))------------------).)).)) ( -16.90) >DroSim_CAF1 95979 76 - 1 GAUGCUCAAUUUGA-UUUUGAUUUAAAUUCCGCUGAGCAUUUUGGUUGAGCUGCAGGUGGG---AUUACUACU------------------GGAACUC ((((((((....((-(((......)))))....))))))))......(((.(.(((.(((.---....)))))------------------).).))) ( -16.90) >DroEre_CAF1 94236 97 - 1 GAUGCUCAAUUCGA-UUUUGCUUUAAAUUCCGAUGAGCAUUUGGGUAGAGCUGUAAUCCCGCCGAUUACUACAGCUUCUGCCCAUUUUUCUGGAGCUC ((((((((..((((-(((......))))..)))))))))))(((((((((((((((((.....)))...)))))).)))))))).............. ( -31.10) >DroYak_CAF1 102912 90 - 1 GAUGUUCAAUUUGA-UUUCGAUUCAAAUUCCGAUGAGCAUUUUGGUAGAGCUGUAGUAGGGCUUA-UACUAGAGCUUUUGCCCAUUUUUCUG------ ((((((((((((((-.......)))))).....))))))))..(((((((((.(((((.......-))))).))).))))))..........------ ( -25.10) >consensus GAUGCUCAAUUUGA_UUUUGAUUUAAAUUCCGCUGAGCAUUUUGGUUGAGCUGCAGUUGGG___AUUACUACAGCUU_UGCCCAUUUUUC_GGAACUC ((((((((((((((........)))))).....))))))))......................................................... ( -8.46 = -8.34 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:42 2006