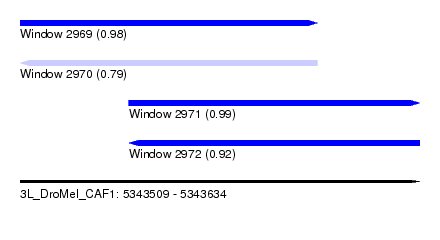
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,343,509 – 5,343,634 |
| Length | 125 |
| Max. P | 0.989535 |

| Location | 5,343,509 – 5,343,602 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -14.32 |
| Energy contribution | -15.52 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
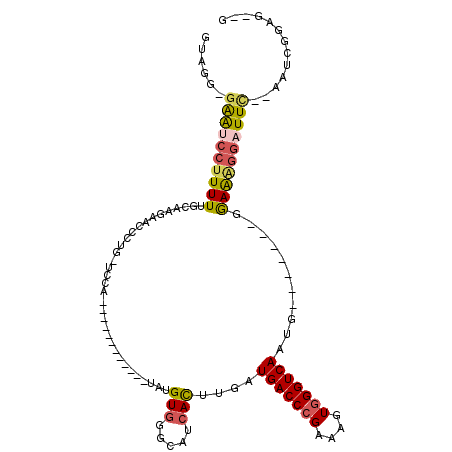
>3L_DroMel_CAF1 5343509 93 + 23771897 AUGGUUACACCUUUGGUUUGCAUUUCCAUUGCUGCCUUCGAUGGAAUCCUUUCCCAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA ..(((..(((...(((.......((((((((.......)))))))).......))).((((((......))))))....)))..)))...... ( -24.64) >DroSec_CAF1 91522 93 + 1 AUGGUUACACCUUUGGUUUGCACUUCCAUUGCUGCCUCCGAUGAAAUUCUUUCCCAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA ..((.....))...(((...(((((.(((((.......)))))..............((((((......))))))..)))))..)))...... ( -20.60) >DroSim_CAF1 93267 93 + 1 AUGGUUACACCUUUGGUUUGAACUUCCAUUGCUGCCUCCGAUUGAAUCCUUUCCCAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA .((((((.(((...))).))).....(((..((..........(((....)))....((((((......))))))...))..))).))).... ( -18.00) >DroEre_CAF1 91433 70 + 1 -----------UUGGGCUUGUAUCUCUUGUG------------GAAUCCACUCCCAUUGACCCACUUUCGGGUCAUCGAGUGAUGCCCACAUC -----------.(((((.....(((.....)------------))...(((((....((((((......))))))..)))))..))))).... ( -25.00) >DroYak_CAF1 100069 93 + 1 GUGACUACGCUUUGGGCUUGUAUCUCAUAUGCUGAUUGCGCUUGAAUCCUUUUCCAUUGACCCACUUUCGGGUCAUCGAGUGUUGCCCACAUC (((....)))..(((((..((((.....)))).....((((((((............((((((......)))))))))))))).))))).... ( -26.40) >consensus AUGGUUACACCUUUGGUUUGCAUUUCCAUUGCUGCCUCCGAUGGAAUCCUUUCCCAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA ..............(((...(((.(((((((.......)))))))............((((((......))))))....)))..)))...... (-14.32 = -15.52 + 1.20)

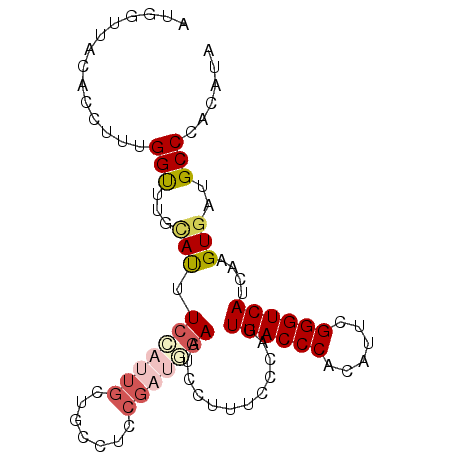
| Location | 5,343,509 – 5,343,602 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5343509 93 - 23771897 UAUGUGGGCAUCACUUGAUGACCCGAAUGUGGGUCAAUGGGAAAGGAUUCCAUCGAAGGCAGCAAUGGAAAUGCAAACCAAAGGUGUAACCAU ...((((....(((((..(((((((....))))))).(((.......((((((.(.......).)))))).......))).)))))...)))) ( -26.14) >DroSec_CAF1 91522 93 - 1 UAUGUGGGCAUCACUUGAUGACCCGAAUGUGGGUCAAUGGGAAAGAAUUUCAUCGGAGGCAGCAAUGGAAGUGCAAACCAAAGGUGUAACCAU ...((((....(((((..(((((((....))))))).(((.......((((((.(.......).)))))).......))).)))))...)))) ( -24.44) >DroSim_CAF1 93267 93 - 1 UAUGUGGGCAUCACUUGAUGACCCGAAUGUGGGUCAAUGGGAAAGGAUUCAAUCGGAGGCAGCAAUGGAAGUUCAAACCAAAGGUGUAACCAU ...((((....(((((..(((((((....))))))).((((((....))).........((....))..........))).)))))...)))) ( -22.00) >DroEre_CAF1 91433 70 - 1 GAUGUGGGCAUCACUCGAUGACCCGAAAGUGGGUCAAUGGGAGUGGAUUC------------CACAAGAGAUACAAGCCCAA----------- ....(((((....((((.(((((((....))))))).)))).((((...)------------)))...........))))).----------- ( -28.00) >DroYak_CAF1 100069 93 - 1 GAUGUGGGCAACACUCGAUGACCCGAAAGUGGGUCAAUGGAAAAGGAUUCAAGCGCAAUCAGCAUAUGAGAUACAAGCCCAAAGCGUAGUCAC (...(((((.....(((.(((((((....))))))).))).......((((...((.....))...))))......)))))...)........ ( -23.90) >consensus UAUGUGGGCAUCACUUGAUGACCCGAAUGUGGGUCAAUGGGAAAGGAUUCAAUCGGAGGCAGCAAUGGAAAUACAAACCAAAGGUGUAACCAU .....((......((((.(((((((....))))))).))))......((((((.(.......).)))))).......)).............. (-13.82 = -14.18 + 0.36)

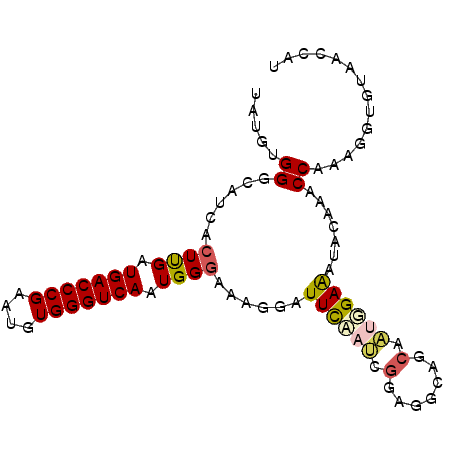
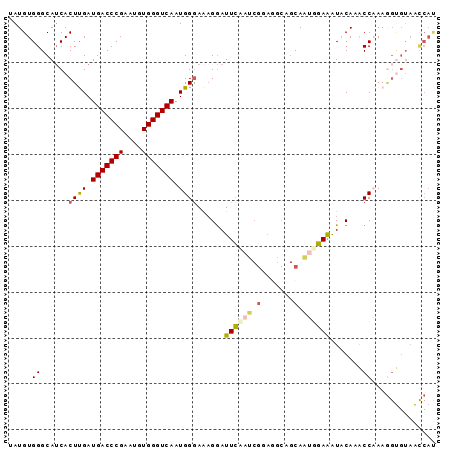
| Location | 5,343,543 – 5,343,634 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.04 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -11.37 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5343543 91 + 23771897 C--CUUCGAUG--GAAUCCUUUCC-------CAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA------------UGGA-CAGGAUUCUUGCAAAAAGGACUC-GCUAC (--(((....(--(((((((.(((-------...((((((......))))))....(((.....)))...------------.)))-.))))))))......))))....-..... ( -29.50) >DroSec_CAF1 91556 91 + 1 C--CUCCGAUG--AAAUUCUUUCC-------CAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA------------UGGA-CAGGGUUCUGGCAAAAAGGCUUC-CCUAC .--.((((...--...........-------...((((((......))))))....(((.....)))...------------))))-.((((....(((......))).)-))).. ( -23.10) >DroSim_CAF1 93301 91 + 1 C--CUCCGAUU--GAAUCCUUUCC-------CAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA------------UGGA-CGGGGUUCUUGCAAAAAGGCUUC-CCUAC .--....(...--(((((((.(((-------...((((((......))))))....(((.....)))...------------.)))-.)))))))...)....(((....-))).. ( -26.40) >DroEre_CAF1 91453 77 + 1 -------------GAAUCCACUCC-------CAUUGACCCACUUUCGGGUCAUCGAGUGAUGCCCACAUC-----------------GAGGUUCCUUGCUAAAAGGAUCC--CUGC -------------.....(((((.-------...((((((......))))))..)))))...........-----------------.(((.(((((.....)))))..)--)).. ( -24.60) >DroYak_CAF1 100103 87 + 1 A--UUGCGCUU--GAAUCCUUUUC-------CAUUGACCCACUUUCGGGUCAUCGAGUGUUGCCCACAUC-----------------GAGGGUCCUUGCUAAAAGGAUCC-CCCAC .--..((((((--((.........-------...((((((......))))))))))))))..........-----------------..((((((((.....))))))))-..... ( -28.40) >DroPer_CAF1 139102 115 + 1 UCUUUCCCAUUACCAAUCGCUUCCCAUUGCGCAUUGACCUAGUUUCGGGUCAUCAAAUGAUGCC-ACAUUGAGUGUCACAUAUGGGGCACGCCCAGUGGAAAAAGGAUCUCAGUUC .............(((((((........))).)))).(((..((((.((.((((....))))))-.(((((.(((((........)))))...))))))))).))).......... ( -28.40) >consensus C__CUCCGAUU__GAAUCCUUUCC_______CAUUGACCCACAUUCGGGUCAUCAAGUGAUGCCCACAUA____________UGGA_CAGGGUCCUUGCAAAAAGGAUCC_CCUAC ...............((((...............((((((......))))))....(((.....)))......................))))((((.....)))).......... (-11.37 = -11.82 + 0.45)

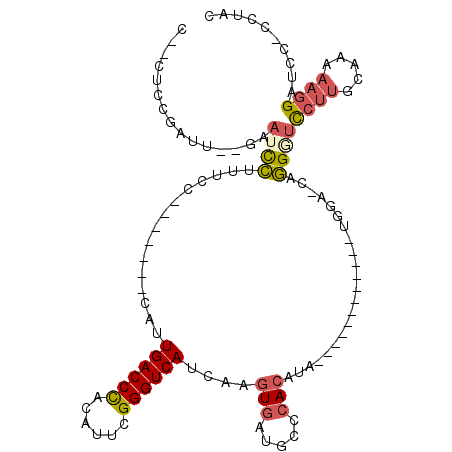
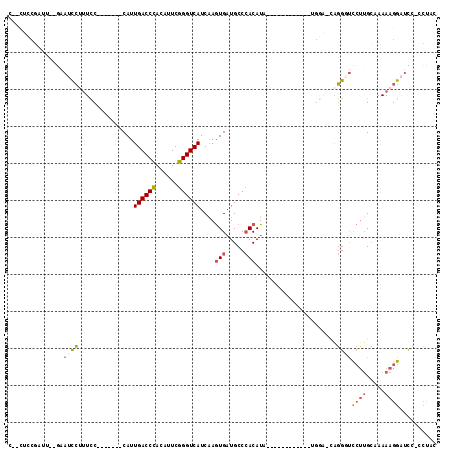
| Location | 5,343,543 – 5,343,634 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.04 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5343543 91 - 23771897 GUAGC-GAGUCCUUUUUGCAAGAAUCCUG-UCCA------------UAUGUGGGCAUCACUUGAUGACCCGAAUGUGGGUCAAUG-------GGAAAGGAUUC--CAUCGAAG--G ...(.-(((((((((((.((((.....((-((((------------....))))))...)))).(((((((....)))))))...-------)))))))))))--).......--. ( -32.10) >DroSec_CAF1 91556 91 - 1 GUAGG-GAAGCCUUUUUGCCAGAACCCUG-UCCA------------UAUGUGGGCAUCACUUGAUGACCCGAAUGUGGGUCAAUG-------GGAAAGAAUUU--CAUCGGAG--G ..(((-....))).....((....(((((-((((------------(((.((((((((....)))).)))).))))))).))..)-------))...((....--..))))..--. ( -27.50) >DroSim_CAF1 93301 91 - 1 GUAGG-GAAGCCUUUUUGCAAGAACCCCG-UCCA------------UAUGUGGGCAUCACUUGAUGACCCGAAUGUGGGUCAAUG-------GGAAAGGAUUC--AAUCGGAG--G .....-(((.(((((..........((((-((((------------(((.((((((((....)))).)))).))))))....)))-------))))))).)))--........--. ( -27.60) >DroEre_CAF1 91453 77 - 1 GCAG--GGAUCCUUUUAGCAAGGAACCUC-----------------GAUGUGGGCAUCACUCGAUGACCCGAAAGUGGGUCAAUG-------GGAGUGGAUUC------------- ...(--((.(((((.....))))).))).-----------------((((....)))).((((.(((((((....))))))).))-------)).........------------- ( -25.90) >DroYak_CAF1 100103 87 - 1 GUGGG-GGAUCCUUUUAGCAAGGACCCUC-----------------GAUGUGGGCAACACUCGAUGACCCGAAAGUGGGUCAAUG-------GAAAAGGAUUC--AAGCGCAA--U (((..-((((((((((..((..(((((((-----------------((.(((.....))))))).....(....).)))))..))-------.))))))))))--...)))..--. ( -33.40) >DroPer_CAF1 139102 115 - 1 GAACUGAGAUCCUUUUUCCACUGGGCGUGCCCCAUAUGUGACACUCAAUGU-GGCAUCAUUUGAUGACCCGAAACUAGGUCAAUGCGCAAUGGGAAGCGAUUGGUAAUGGGAAAGA ...........((((..(.((..(.(((..(((((.((((.(((.....))-)...........(((((........)))))...)))))))))..))).)..))...)..)))). ( -32.20) >consensus GUAGG_GAAUCCUUUUUGCAAGAACCCUG_UCCA____________UAUGUGGGCAUCACUUGAUGACCCGAAAGUGGGUCAAUG_______GGAAAGGAUUC__AAUCGGAG__G ......((((((((((.................................(((.....)))....(((((((....)))))))...........))))))))))............. (-16.09 = -16.40 + 0.31)
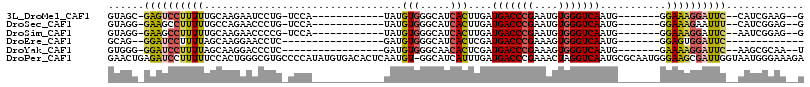

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:38 2006