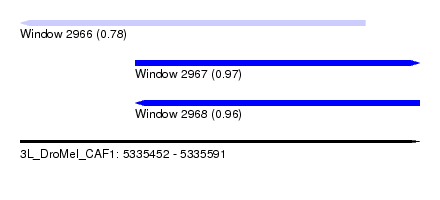
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,335,452 – 5,335,591 |
| Length | 139 |
| Max. P | 0.973716 |

| Location | 5,335,452 – 5,335,572 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -39.68 |
| Energy contribution | -40.35 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
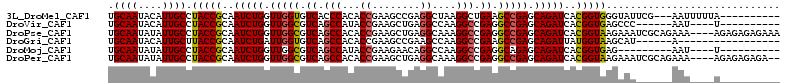
>3L_DroMel_CAF1 5335452 120 - 23771897 CGUGAUCUGCUCGGCUUCAGCCUUAGCCUCGGCUUCGGUGUGGGUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUA ((..(((((...(((....)))..(((....)))..(((((.....)))))...)))))..))....((((....))))..((((((((((((((.......)))))))).)).)))).. ( -43.20) >DroPse_CAF1 129524 120 - 1 CGUGAUCUGCUCGGCCUCGGCCUUUGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCAUCGAAUGGCAGGCGGAGCACACCUGCUUGUACACCUCAUA .(..(((((...(((.((((((..((((........)))).)))))).)))...)))))..)(((..((((....))))........((((((((.......))))))))..)))..... ( -45.20) >DroGri_CAF1 89969 120 - 1 CAUAAUCUGCUCGGCUUCGGCCUUGGCUUCGGCUUCGGUGUGGCUGACACCAAUCAGAUUGCGGUAAGCAAUGUAUUGCAAGGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUA ...((((((...(((....)))((((..((((((.......))))))..)))).))))))((((((.......))))))..(..(((((((((((.......)))))))).)))..)... ( -42.80) >DroWil_CAF1 87998 120 - 1 CGUAAUCUGCUCGGCUUCCGCCUUAGCCUCAGCUUCGGUAUGGGUAACGCCAACCAAGUUACGGUAGGCGAUAUAUUGCAUCGAGUGGCAGGCGGAGCAGACCUGCUUGUAGACCUCAUA .(((.(((((((.(((((((((...((((..((....))..))))..((((.(((.......))).))))............).)))).)))).)))))))..))).............. ( -43.70) >DroMoj_CAF1 19312 120 - 1 CGUGAUCUGCUCUGCCUCGGCCUUGGCCUGUUCUUCGGUAUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCAUCGAGUGGCAAGCAGAGCACACCUGCUUGUACACCUCAUA ((..(((((....((.((((((...(((........)))..)))))).))....)))))..))....((((....))))...(((((((((((((.......)))))))).)).)))... ( -46.60) >DroPer_CAF1 129908 120 - 1 CGUGAUCUGCUCGGCCUCGGCCUUUGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCAUCGAAUGGCAGGCGGAGCACACCUGCUUGUACACCUCAUA .(..(((((...(((.((((((..((((........)))).)))))).)))...)))))..)(((..((((....))))........((((((((.......))))))))..)))..... ( -45.20) >consensus CGUGAUCUGCUCGGCCUCGGCCUUAGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCAUA (((((((((...(((.((((((...(((........)))..)))))).)))...)))))))))....((((....))))...(((..((((((((.......))))))))....)))... (-39.68 = -40.35 + 0.67)

| Location | 5,335,492 – 5,335,591 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -30.94 |
| Energy contribution | -30.83 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5335492 99 + 23771897 UGCAAUACAUUGCCUACCGCAAUCUGGUUGGUGUCACCCACACCGAAGCCGAGGCUAAGGCUGAAGCCGAGCAGAUCACGGUGGGUAUUCG---AAUUUUUA---------- ..........(((((((((..(((((.(((((.(((......((..(((....)))..)).))).))))).)))))..)))))))))....---........---------- ( -36.40) >DroVir_CAF1 109069 92 + 1 UGCAAUACAUUGCCUACCGCAAUCUGGUUGGCGUCAGCCAUACCGAAGCUGAGGCCAAGGCCGAGGCCGAGCAGAUCACGGUGAGCCC------AAU----U---------- ...........((.(((((..(((((.(((((.(((((.........))))).)))))(((....)))...)))))..))))).))..------...----.---------- ( -36.90) >DroPse_CAF1 129564 108 + 1 UGCAAUAUAUUGCCUACCGCAAUCUGGUUGGCGUCAGCCACACCGAAGCUGAGGCAAAGGCCGAGGCCGAGCAGAUCACGGUAAGAAAUCGCAGAAA----AGAGAGAGAAA .((((....)))).(((((..(((((.(((((.((.(((...((........))....))).)).))))).)))))..)))))..............----........... ( -35.10) >DroGri_CAF1 90009 89 + 1 UGCAAUACAUUGCUUACCGCAAUCUGAUUGGUGUCAGCCACACCGAAGCCGAAGCCAAGGCCGAAGCCGAGCAGAUUAUGGUAAGCAU------A----------------- ..........(((((((((.((((((.(((((.((.(((...................))).)).))))).)))))).))))))))).------.----------------- ( -32.61) >DroMoj_CAF1 19352 89 + 1 UGCAAUAUAUUGCCUACCGCAAUCUGGUUGGCGUCAGCCAUACCGAAGAACAGGCCAAGGCCGAGGCAGAGCAGAUCACGGUGAG---------AAU----U---------- .((((....)))).(((((..(((((.((.((.((.(((...((........))....))).)).)).)).)))))..)))))..---------...----.---------- ( -29.00) >DroPer_CAF1 129948 106 + 1 UGCAAUAUAUUGCCUACCGCAAUCUGGUUGGCGUCAGCCACACCGAAGCUGAGGCAAAGGCCGAGGCCGAGCAGAUCACGGUAAGAAAUCGCAGAAA----AGAGAGAGA-- .((((....)))).(((((..(((((.(((((.((.(((...((........))....))).)).))))).)))))..)))))..............----.........-- ( -35.10) >consensus UGCAAUACAUUGCCUACCGCAAUCUGGUUGGCGUCAGCCACACCGAAGCUGAGGCCAAGGCCGAGGCCGAGCAGAUCACGGUAAGAAA______AAU____A__________ .((((....)))).(((((..(((((.(((((.((.(((...((........))....))).)).))))).)))))..)))))............................. (-30.94 = -30.83 + -0.11)

| Location | 5,335,492 – 5,335,591 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -34.10 |
| Energy contribution | -34.52 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5335492 99 - 23771897 ----------UAAAAAUU---CGAAUACCCACCGUGAUCUGCUCGGCUUCAGCCUUAGCCUCGGCUUCGGUGUGGGUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCA ----------........---..(((((((((((..(((((...(((....)))..(((....)))..(((((.....)))))...)))))..)))).))....)))))... ( -33.10) >DroVir_CAF1 109069 92 - 1 ----------A----AUU------GGGCUCACCGUGAUCUGCUCGGCCUCGGCCUUGGCCUCAGCUUCGGUAUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUGUAUUGCA ----------.----...------..((..((((..(((((...(((....)))(((((.((((((.......)))))).))))).)))))..))))..))........... ( -42.60) >DroPse_CAF1 129564 108 - 1 UUUCUCUCUCU----UUUCUGCGAUUUCUUACCGUGAUCUGCUCGGCCUCGGCCUUUGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCA ...........----.....(((((..(((((((..(((((...(((.((((((..((((........)))).)))))).)))...)))))..)))))))......))))). ( -43.60) >DroGri_CAF1 90009 89 - 1 -----------------U------AUGCUUACCAUAAUCUGCUCGGCUUCGGCCUUGGCUUCGGCUUCGGUGUGGCUGACACCAAUCAGAUUGCGGUAAGCAAUGUAUUGCA -----------------.------.((((((((.(((((((...(((....)))((((..((((((.......))))))..)))).))))))).)))))))).......... ( -38.90) >DroMoj_CAF1 19352 89 - 1 ----------A----AUU---------CUCACCGUGAUCUGCUCUGCCUCGGCCUUGGCCUGUUCUUCGGUAUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCA ----------.----...---------((.((((..(((((....((.((((((...(((........)))..)))))).))....)))))..))))))((((....)))). ( -36.30) >DroPer_CAF1 129948 106 - 1 --UCUCUCUCU----UUUCUGCGAUUUCUUACCGUGAUCUGCUCGGCCUCGGCCUUUGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCA --.........----.....(((((..(((((((..(((((...(((.((((((..((((........)))).)))))).)))...)))))..)))))))......))))). ( -43.60) >consensus __________U____AUU______AUGCUCACCGUGAUCUGCUCGGCCUCGGCCUUGGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCA ..............................(((((((((((...(((.((((((...(((........)))..)))))).)))...)))))))))))..((((....)))). (-34.10 = -34.52 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:35 2006