
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,328,691 – 5,328,785 |
| Length | 94 |
| Max. P | 0.760856 |

| Location | 5,328,691 – 5,328,785 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -14.61 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5328691 94 - 23771897 UAAACCUGCGCCUUUUCAAUGUAGCCCUUCAUGGAUAAUUAACACGUGAGCGUUGUGUGACUAUAUAGCAAA--AUCGUGUUAAA-------AUCG----UGUUCGC--- .......((((..............((.....))....((((((((.....(((((((....)))))))...--..)))))))).-------...)----)))....--- ( -17.10) >DroVir_CAF1 102876 99 - 1 --AUCCAUUGCGGUUCCAAUGUAGCCCCUCAUGGUUAAUUAACACGAGCCCGUUGUGUGACUAUAUAACAGU--UUCGUGUUAAA-------ACAAAAAGUGUACAAAAA --...(((((......)))))(((((......))))).((((((((((.(.(((((((....))))))).).--)))))))))).-------.................. ( -22.60) >DroGri_CAF1 84310 90 - 1 --UUCCAGUGCAUUUCCAAUGUAGCCCUUUAUGGUUAAUUAGCACGAGCCCGUUGUGUGACUAUAUAACAGU--UUCGUGUUAAA-------UCUC---------AAAAA --...................(((((......))))).((((((((((.(.(((((((....))))))).).--)))))))))).-------....---------..... ( -20.20) >DroMoj_CAF1 12994 95 - 1 --ACCCAGUGCGGUUCCAAUGUAGCCCUUCAUGGUUAAUUAACACGAGCUCGUUGUGUGACUAUAUCGCUGU--UUCGUGUUAAA-------ACUAUAUCU----AUAAA --.....(((..(((......(((((......))))).(((((((((((.((.((((....)))).))..).--)))))))))))-------))..)))..----..... ( -17.50) >DroAna_CAF1 74687 96 - 1 UUAAUUAGUGCGUUUCCAAUGUAGCCCCUCAUGGAUAAUUAACACGAGAUCGUUGUGUGACUAUAUAGCAGU--AUCGUGUUAAA-------ACAAAA--AUUUCGC--- .........(((..((((.((........))))))...(((((((((.((.(((((((....))))))).))--.))))))))).-------......--....)))--- ( -21.40) >DroPer_CAF1 122257 101 - 1 C--ACCAGUGCUUUUCCAAUGUAGCCCUUCACGCUUGAUUAACACGAGAGCGUUGUGUGACAAUAUAGCAGUUUUUCGUGUUAAAUACCCAAAUAU----CGUACAA--- .--....((((...........(((.......)))...((((((((((((((((((((....))))))).).))))))))))))............----.))))..--- ( -23.30) >consensus __AACCAGUGCGUUUCCAAUGUAGCCCUUCAUGGUUAAUUAACACGAGACCGUUGUGUGACUAUAUAGCAGU__UUCGUGUUAAA_______ACAA____UGU_CAA___ .....................(((((......))))).((((((((((...(((((((....))))))).....)))))))))).......................... (-14.61 = -15.28 + 0.67)

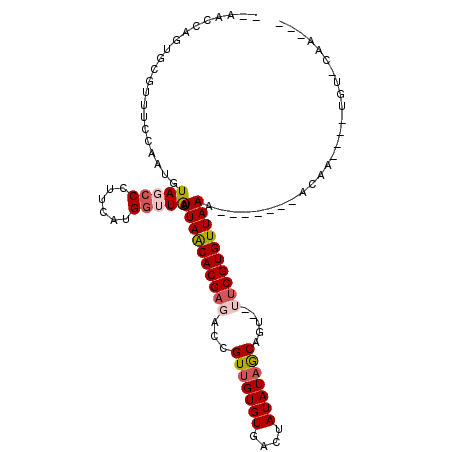
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:29 2006