
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,325,553 – 5,325,676 |
| Length | 123 |
| Max. P | 0.996705 |

| Location | 5,325,553 – 5,325,651 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -20.49 |
| Consensus MFE | -13.21 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5325553 98 - 23771897 AUAAUUU-UCCCAAAUUA-----CCAAGUAGGAAAAUUUACCACAUACCCGUGAAUUUUUCAG--CUAGCGGGUAUGAAAAUUUCGCACGAUUUUCCCGCCAUCAU .......-..........-----....(..((((((.......(((((((((...........--...)))))))))......((....))))))))..)...... ( -19.94) >DroSec_CAF1 72813 98 - 1 AUAAUUU-UCCCAAAUUA-----CCAAGUGGGAAAAUUUACCACAUACCCGUGAACUUUUCAG--CUAGCGGGUAUGAAAAUUUCACACGAUUUUCCCGCCAUCGU .......-..........-----....(((((((((.......(((((((((...((....))--...)))))))))......((....)))))))))))...... ( -25.80) >DroSim_CAF1 74821 98 - 1 AUAAUUU-UUCCAAAUUA-----CCAAGUGGGAAAAUUUACCACAUACCCGUGAACUUUUCAG--CUAGCGGGUAUGAAAAUUUCACACGAUUUUCCCGCCAUCGU .......-..........-----....(((((((((.......(((((((((...((....))--...)))))))))......((....)))))))))))...... ( -25.80) >DroEre_CAF1 72737 98 - 1 AUAAUUU-UCCCAAAUUA-----UCGAAUAGGAAAAUUUACCGCAUACCCGUACACUUUUCUG--CUAGCGGGUAUGAACAUUUCGGACGCUUUUCCCGCCAUCGU (((((((-....))))))-----)(((...((((((....((((((((((((...(......)--...))))))))).......)))....)))))).....))). ( -20.41) >DroYak_CAF1 81371 98 - 1 CUAAUUU-UCCCAAAUUA-----UCAAAUAAGAAAAUUUACCGCAUACCCGUCAACUUUUCAG--CUAGCGGGUAUGAAAAUUUCGCACGAUUUUCCCGCCAUCGU .......-....(((((.-----((......)).)))))...(((((((((..(.((....))--.)..)))))))(((((((......)))))))..))...... ( -16.60) >DroAna_CAF1 71697 103 - 1 UUAUUUUUUCCCACCUUUCACCUUCAGAUA--CAAAUACAACAUAUACCCGUGAUAUUUUCGGGAAUAGCGGGUAUGAAAAUUU-GCACGAUUUUUCCGCUUUCUU .......(((((.....((.......))..--.(((((..((........))..)))))..))))).(((((....(((((((.-....))))))))))))..... ( -14.40) >consensus AUAAUUU_UCCCAAAUUA_____CCAAAUAGGAAAAUUUACCACAUACCCGUGAACUUUUCAG__CUAGCGGGUAUGAAAAUUUCGCACGAUUUUCCCGCCAUCGU ..............................((((((((.....(((((((((................)))))))))............))))))))......... (-13.21 = -14.07 + 0.86)



| Location | 5,325,579 – 5,325,676 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.95 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5325579 97 + 23771897 UUUCAUACCCGCUAG--CUGAAAAAUUCACGGGUAUGUGGUAAAUUUUCCUACUUGG-----UAAUUUGGGA-AAAUUAUCU-------------UUGCUAUCUUAAUCAAAUAAAUU-- ...((((((((....--.(((.....)))))))))))(((((((((((((((..(..-----..)..)))))-)))).....-------------)))))).................-- ( -20.90) >DroSec_CAF1 72839 93 + 1 UUUCAUACCCGCUAG--CUGAAAAGUUCACGGGUAUGUGGUAAAUUUUCCCACUUGG-----UAAUUUGGGA-AAAUUAUCU-------------UAGCUAUCUUAAUCGAAUA------ ...((((((((..((--((....))))..)))))))).(((((.((((((((..(..-----..)..)))))-)))))))).-------------...................------ ( -29.10) >DroSim_CAF1 74847 97 + 1 UUUCAUACCCGCUAG--CUGAAAAGUUCACGGGUAUGUGGUAAAUUUUCCCACUUGG-----UAAUUUGGAA-AAAUUAUCU-------------UAGCUAUCUUAAUCAAAUAUCUU-- ....(((((((..((--((....))))..)))))))((((.........))))..((-----((((((....-)))))))).-------------.......................-- ( -24.90) >DroEre_CAF1 72763 97 + 1 GUUCAUACCCGCUAG--CAGAAAAGUGUACGGGUAUGCGGUAAAUUUUCCUAUUCGA-----UAAUUUGGGA-AAAUUAUCU-------------GUUCCAUCUUAAUCAAAUAAAUU-- ...((((((((...(--(......))...))))))))(((..((((((((((.....-----.....)))))-)))))..))-------------)......................-- ( -23.50) >DroYak_CAF1 81397 99 + 1 UUUCAUACCCGCUAG--CUGAAAAGUUGACGGGUAUGCGGUAAAUUUUCUUAUUUGA-----UAAUUUGGGA-AAAUUAGCU-------------GUGCAAUCUUAAUCAUGUGAAUUAU .((((((((((.(((--((....))))).)))))))(((((.((((((((((..(..-----..)..)))))-))))).)))-------------))................))).... ( -30.30) >DroAna_CAF1 71722 111 + 1 UUUCAUACCCGCUAUUCCCGAAAAUAUCACGGGUAUAUGUUGUAUUUG--UAUCUGAAGGUGAAAGGUGGGAAAAAAUAAGUGUUCCCAUUAUUUUAACAACCUUUAU-----GUUUU-- ...((((((((((.(((((((.....)).((((((((.........))--))))))..)).))).))))))..(((((((.((....))))))))).........)))-----)....-- ( -23.50) >consensus UUUCAUACCCGCUAG__CUGAAAAGUUCACGGGUAUGUGGUAAAUUUUCCUACUUGA_____UAAUUUGGGA_AAAUUAUCU_____________UAGCUAUCUUAAUCAAAUAAAUU__ ...((((((((..................)))))))).(((((.((.(((((...............))))).)).)))))....................................... (-12.86 = -13.06 + 0.20)

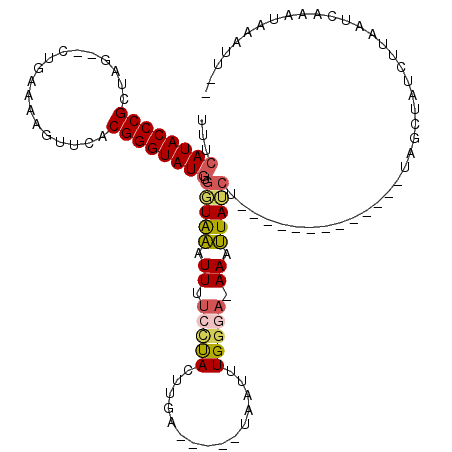
| Location | 5,325,579 – 5,325,676 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.95 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -13.13 |
| Energy contribution | -12.99 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5325579 97 - 23771897 --AAUUUAUUUGAUUAAGAUAGCAA-------------AGAUAAUUU-UCCCAAAUUA-----CCAAGUAGGAAAAUUUACCACAUACCCGUGAAUUUUUCAG--CUAGCGGGUAUGAAA --...((((((.....))))))...-------------....(((((-(((.......-----.......)))))))).....(((((((((...........--...)))))))))... ( -18.98) >DroSec_CAF1 72839 93 - 1 ------UAUUCGAUUAAGAUAGCUA-------------AGAUAAUUU-UCCCAAAUUA-----CCAAGUGGGAAAAUUUACCACAUACCCGUGAACUUUUCAG--CUAGCGGGUAUGAAA ------...................-------------....(((((-(((((.....-----.....)))))))))).....(((((((((...((....))--...)))))))))... ( -24.20) >DroSim_CAF1 74847 97 - 1 --AAGAUAUUUGAUUAAGAUAGCUA-------------AGAUAAUUU-UUCCAAAUUA-----CCAAGUGGGAAAAUUUACCACAUACCCGUGAACUUUUCAG--CUAGCGGGUAUGAAA --.((.(((((.....))))).)).-------------....(((((-(((((.....-----.....)))))))))).....(((((((((...((....))--...)))))))))... ( -22.90) >DroEre_CAF1 72763 97 - 1 --AAUUUAUUUGAUUAAGAUGGAAC-------------AGAUAAUUU-UCCCAAAUUA-----UCGAAUAGGAAAAUUUACCGCAUACCCGUACACUUUUCUG--CUAGCGGGUAUGAAC --..((((((((((......((((.-------------........)-)))......)-----)))))))))...........(((((((((...(......)--...)))))))))... ( -22.30) >DroYak_CAF1 81397 99 - 1 AUAAUUCACAUGAUUAAGAUUGCAC-------------AGCUAAUUU-UCCCAAAUUA-----UCAAAUAAGAAAAUUUACCGCAUACCCGUCAACUUUUCAG--CUAGCGGGUAUGAAA ...............((((((((..-------------.)).)))))-)...(((((.-----((......)).)))))....((((((((..(.((....))--.)..))))))))... ( -15.20) >DroAna_CAF1 71722 111 - 1 --AAAAC-----AUAAAGGUUGUUAAAAUAAUGGGAACACUUAUUUUUUCCCACCUUUCACCUUCAGAUA--CAAAUACAACAUAUACCCGUGAUAUUUUCGGGAAUAGCGGGUAUGAAA --.....-----..((((((.(..(((((((((....)).)))))))..)..))))))............--...........(((((((((..(((((....))))))))))))))... ( -23.10) >consensus __AAUUUAUUUGAUUAAGAUAGCAA_____________AGAUAAUUU_UCCCAAAUUA_____CCAAAUAGGAAAAUUUACCACAUACCCGUGAACUUUUCAG__CUAGCGGGUAUGAAA ...................................................................................(((((((((................)))))))))... (-13.13 = -12.99 + -0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:27 2006