
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
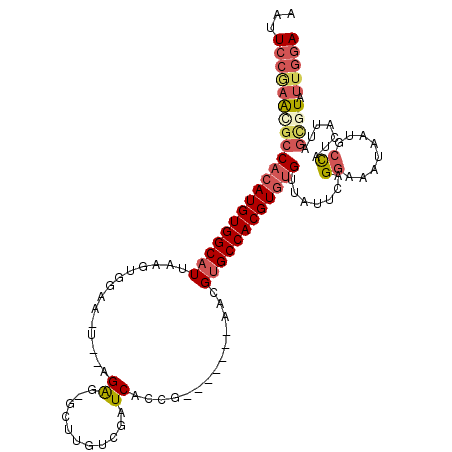
| Location | 5,323,669 – 5,323,775 |
| Length | 106 |
| Max. P | 0.831220 |

| Location | 5,323,669 – 5,323,775 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -18.62 |
| Energy contribution | -19.91 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5323669 106 + 23771897 AAUUCCGAACGCCACAUGUGGCAUUAAGUGGAA-UAUAGAG-GCAUGUCGAUCACCG-------AACGUGCCACGUGUGUUAUUCAGGAAAUAAUGCCAUCAUUAGCAUAUUGGA ...(((((..((...(((((((((((....(((-((..(.(-((((((((.....))-------.))))))).)......)))))......)))))))).)))..))...))))) ( -31.00) >DroSec_CAF1 70940 104 + 1 AAUUCCGAACACCACAUGUGGCAUUAAGUGGAA-U--AGAG-GCUUGUCUAUCACCG-------AACGUGCCACGUGUGUUAUUCCGGAAAUAAUGCCAUCAUUAGCGUAUUGGA ..(((((.....((((((((((((....(((.(-(--(((.-.....)))))..)))-------...))))))))))))......))))).(((((((.......).)))))).. ( -32.90) >DroSim_CAF1 72902 104 + 1 AAUUCCGAACGCCACAUGUGGCAUUAAGUGGAA-U--AGAG-GCUUGUCGAUCACCG-------AACGUGCCACGUGUGUUAUUCCGGAAAUAAUGCCAUCAUUAGCGUAUUGGA ...(((((((((...(((((((((((..(((((-(--((.(-((.(((((.....))-------.))).))).......))))))))....)))))))).)))..)))).))))) ( -33.80) >DroEre_CAF1 70881 105 + 1 AAUUCCGAAUGCCACAUGUGGCAUUAAGUAGAA-U--AGAGAGCUAUUCGAUCACUG-------AACGCGCCACGUGUGUUAUUAAGGAAAUAAUUCCAUUAUAAGCGUAUUGGA ...(((((((((((((((((((....(((.(((-(--((....))))))....))).-------.....)))))))))).......((((....)))).......)))).))))) ( -33.90) >DroYak_CAF1 79454 105 + 1 AAUUCCAAACGCCACAUGUGGCAUUAACUAGAA-U--AGAGUGCUUUUCGAUCACUG-------AACGUGCCACGUGUGUUAUGCAGGAAAUAAUGCCAUUAUUAGCGUAUUGGA ...(((((((((((((((((((((....(((..-.--.(((.....))).....)))-------...))))))))))))....(((........)))........)))).))))) ( -32.00) >DroAna_CAF1 69990 112 + 1 AAUUCGUAGCGCCAAAUGUGGCAUUUGAUGAAAAUGUGGGG-GCUCGACGAUCGGUGUCUUCUUCACGUGCCACGUGAGUGAUCCAGGAAAUAAAUAUAU--UCAAUAUAGUGAA ..(((....(((...(((((((((.(((.........((((-(((((.....))).)))))).))).)))))))))..))).....)))..........(--(((......)))) ( -26.90) >consensus AAUUCCGAACGCCACAUGUGGCAUUAAGUGGAA_U__AGAG_GCUUGUCGAUCACCG_______AACGUGCCACGUGUGUUAUUCAGGAAAUAAUGCCAUCAUUAGCGUAUUGGA ...(((((((((((((((((((((..............((...........))..............)))))))))))).......((........)).......)))).))))) (-18.62 = -19.91 + 1.28)


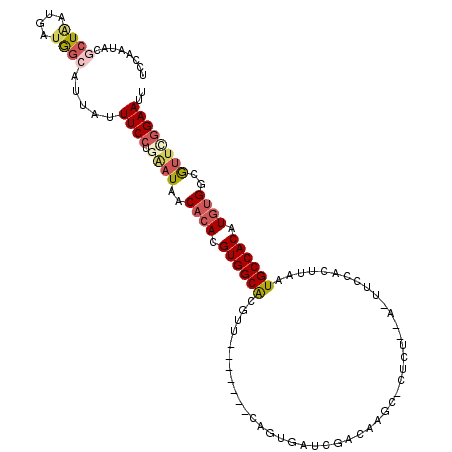
| Location | 5,323,669 – 5,323,775 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -14.93 |
| Energy contribution | -16.05 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5323669 106 - 23771897 UCCAAUAUGCUAAUGAUGGCAUUAUUUCCUGAAUAACACACGUGGCACGUU-------CGGUGAUCGACAUGC-CUCUAUA-UUCCACUUAAUGCCACAUGUGGCGUUCGGAAUU (((...(((((((((.((((((((......(((((((....))((((.((.-------((.....)))).)))-)....))-)))....))))))))))).))))))..)))... ( -30.40) >DroSec_CAF1 70940 104 - 1 UCCAAUACGCUAAUGAUGGCAUUAUUUCCGGAAUAACACACGUGGCACGUU-------CGGUGAUAGACAAGC-CUCU--A-UUCCACUUAAUGCCACAUGUGGUGUUCGGAAUU ........((((....)))).....(((((((....((((.((((((....-------..(..(((((.....-.)))--)-)..)......)))))).))))...))))))).. ( -32.80) >DroSim_CAF1 72902 104 - 1 UCCAAUACGCUAAUGAUGGCAUUAUUUCCGGAAUAACACACGUGGCACGUU-------CGGUGAUCGACAAGC-CUCU--A-UUCCACUUAAUGCCACAUGUGGCGUUCGGAAUU (((...(((((((((.((((((((.....((((((((....))(((..((.-------((.....))))..))-)..)--)-))))...))))))))))).))))))..)))... ( -32.90) >DroEre_CAF1 70881 105 - 1 UCCAAUACGCUUAUAAUGGAAUUAUUUCCUUAAUAACACACGUGGCGCGUU-------CAGUGAUCGAAUAGCUCUCU--A-UUCUACUUAAUGCCACAUGUGGCAUUCGGAAUU (((.....(.((((...((((....))))...))))).((((((..(((((-------.((((...((((((....))--)-))))))).)))))..))))))......)))... ( -27.20) >DroYak_CAF1 79454 105 - 1 UCCAAUACGCUAAUAAUGGCAUUAUUUCCUGCAUAACACACGUGGCACGUU-------CAGUGAUCGAAAAGCACUCU--A-UUCUAGUUAAUGCCACAUGUGGCGUUUGGAAUU (((((.((((((....((((((((.....(((...((....)).)))....-------.((((.........))))..--.-.......))))))))....)))))))))))... ( -28.20) >DroAna_CAF1 69990 112 - 1 UUCACUAUAUUGA--AUAUAUUUAUUUCCUGGAUCACUCACGUGGCACGUGAAGAAGACACCGAUCGUCGAGC-CCCCACAUUUUCAUCAAAUGCCACAUUUGGCGCUACGAAUU ....(((....((--(((....)))))..)))........((((((..(((((((.(((.......)))(...-...)...))))))).....((((....)))))))))).... ( -20.40) >consensus UCCAAUACGCUAAUGAUGGCAUUAUUUCCUGAAUAACACACGUGGCACGUU_______CAGUGAUCGACAAGC_CUCU__A_UUCCACUUAAUGCCACAUGUGGCGUUCGGAAUU ........((((....)))).....((((.((((..((((.((((((.............................................)))))).))))..)))))))).. (-14.93 = -16.05 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:25 2006