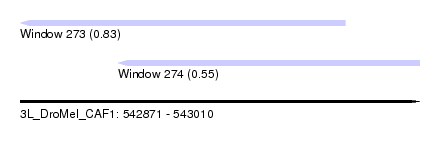
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 542,871 – 543,010 |
| Length | 139 |
| Max. P | 0.831022 |

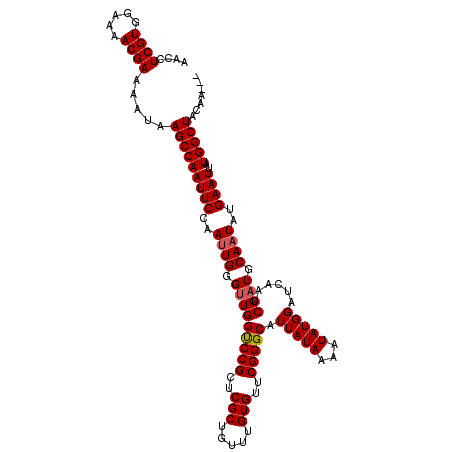
| Location | 542,871 – 542,984 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 97.55 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -27.56 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.831022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 542871 113 - 23771897 AACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAUGGAUCAAUCGAUGCAAUAUGAAUUAUGGCUACAA--- ....((((.....)))).....(((((((((..((((.(((((((((..(((.....)))..))))).(((((...)))))......)))).))))..))))..)))))....--- ( -28.20) >DroSec_CAF1 16886 113 - 1 AACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAAUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAUGGAUCAAUCGAUGCAAUAUGAAUUAUGGCUACAA--- ....((((.....)))).....(((((((((...((.((((((((((((((...........)))))(((....)))..))))))))).)).......))))..)))))....--- ( -27.50) >DroSim_CAF1 17795 113 - 1 AACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGCCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAUGGAUCAAUCGAUGCAAUAUGAAUUAUGGCUACAA--- ....((((.....)))).....(((((((((..((((.(((((((((..(((.....)))..))))).(((((...)))))......)))).))))..))))..)))))....--- ( -30.90) >DroEre_CAF1 17266 113 - 1 AACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGCUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAUGGAUCAAUCGAUGCAAUAUGAAUUAUGGCUACAA--- ....((((.....)))).....(((((((((...........(((((..(((.....)))..)))))...........((.(((....))).))....))))..)))))....--- ( -25.90) >DroYak_CAF1 17497 116 - 1 AACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAUGGAUCAAUCGAUGCAAUAUGAAUUAUGGCUACGAGCA ...(((((((..........(((.(((((....))))).)))(((((..(((.....)))..))))).((((((.(((((.(((....))).).))))...))))))))))))).. ( -30.50) >consensus AACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAUGGAUCAAUCGAUGCAAUAUGAAUUAUGGCUACAA___ ....((((.....)))).....(((((((((..((((.(((((((((..(((.....)))..))))).(((((...)))))......)))).))))..))))..)))))....... (-27.56 = -27.80 + 0.24)

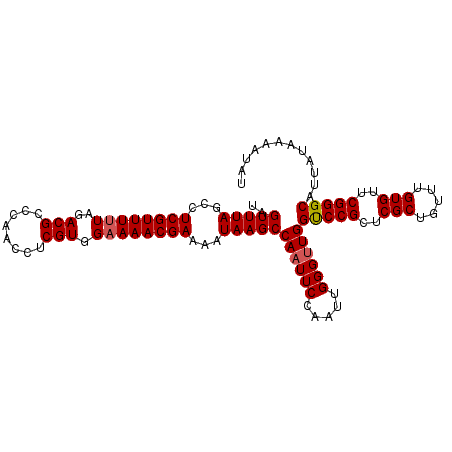
| Location | 542,905 – 543,010 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 98.48 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 542905 105 - 23771897 UAGUUUAGCCUCGUUUUUAGACGCCCAACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAU ..(((((...((((((((..(((........))).))))))))...)))))((((((.....))))))(((((..(((.....)))..)))))............ ( -26.80) >DroSec_CAF1 16920 105 - 1 UAGUUUAGCCUCGUUUUUAGACGCCCAACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAAUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAU ..(((((...((((((((..(((........))).))))))))...)))))((((((.....))))))(((((..(((.....)))..)))))............ ( -26.80) >DroSim_CAF1 17829 105 - 1 UAGUUUAGCCUCGUUUUUAGACGCCCAACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGCCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAU ..(((((...((((((((..(((........))).))))))))...)))))((((((.....))))))(((((..(((.....)))..)))))............ ( -29.50) >DroEre_CAF1 17300 105 - 1 UAGUUUAGCCUCGUUUUUAGACGCCCAACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGCUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAU .....(((((((((((((..(((........))).))))))))........((((....)))))))))(((((..(((.....)))..)))))............ ( -27.30) >DroYak_CAF1 17534 105 - 1 UAGUUUAGCCUCGUUUCUAGACGCCCAACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAU ..((((((........))))))...((((((..(((((................)))))...))))))(((((..(((.....)))..)))))............ ( -27.29) >consensus UAGUUUAGCCUCGUUUUUAGACGCCCAACCUCGUGGAAAACGAAAAUAAGCCAAUUCCAAUUGGGUUGGUCCGCUCGCUGUUUGUGUUCGGGCAUUAUAAAAUAU ..(((((...((((((((..(((........))).))))))))...)))))((((((.....))))))(((((..(((.....)))..)))))............ (-26.20 = -26.44 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:22 2006