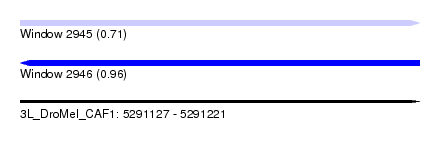
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,291,127 – 5,291,221 |
| Length | 94 |
| Max. P | 0.955630 |

| Location | 5,291,127 – 5,291,221 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -15.69 |
| Energy contribution | -16.97 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
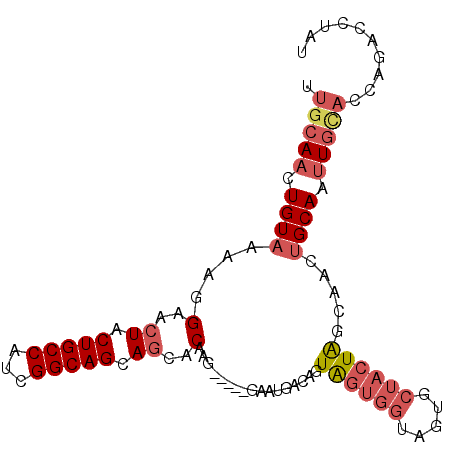
>3L_DroMel_CAF1 5291127 94 + 23771897 UUGCAACUGUAAAAGGAACUACUGCCAUCGGCAGCAGCACAAG-----GAAUGACAGUAGUGGUAGUGCUACUAGCAACUGCAAUUGCAGCAAACCUAU ((((.(((((.....(..((.(((((...))))).))..)...-----.....)))))(((((.....))))).))))((((....))))......... ( -24.12) >DroSec_CAF1 38546 94 + 1 UUGCAACUGUAAAAGGAACUACUGCCAUCGGCAGCAGCACAAG-----GAAUGACAGUAGUGGCAGUGCUACUAGCAACUGCAGUUGCACCAGACCUAU .((((((((((.......((.(((((...))))).))......-----...((..((((((......))))))..))..)))))))))).......... ( -29.00) >DroSim_CAF1 39373 94 + 1 UUGCAACUGUAAAAGGAACUACUGCCAUCGGCAGCAGCGCAAG-----GAAUGACAGUAGUGGUAGUGCUACUAGCAACUGCAGUUGCACCAGACCUAU .((((((((((....(..((.(((((...))))).))..)...-----...((..((((((......))))))..))..)))))))))).......... ( -29.90) >DroEre_CAF1 38106 81 + 1 UUGCAACUGUAAAAGGGACUACUGCCAUCGGCAGCAGCACCAG-----GAAUGACAGUAGUGGUAGUGCUACUAGCAACUGCACAU------------- ..((.(((((....((..((.(((((...))))).))..))..-----.....))))).)).(((((((.....)).)))))....------------- ( -24.30) >DroYak_CAF1 39864 94 + 1 UUGCAACUGUAAGAGGGACUACUGCCAUCGGCAGCAGCACAAG-----GAAUGACAGUGGUGGUUGUACUACUAGCAACUGCAAUUGCAGAAGACCUAU ((((.(((((.....(..((.(((((...))))).))..)...-----.....)))))(((((.....))))).))))((((....))))......... ( -26.52) >DroPer_CAF1 79141 88 + 1 UUGCAACUGUAAAAGGGGCUACUGCCAUCGGCAGCAAUACCAGCAGCAGAAUGGCUGUAGCAGCAACAAGAAUGG----GGCAAUUGUACUG------- .(((((...........(((((.((((((.((.((.......)).)).).))))).))))).((..((....)).----.))..)))))...------- ( -25.80) >consensus UUGCAACUGUAAAAGGAACUACUGCCAUCGGCAGCAGCACAAG_____GAAUGACAGUAGUGGUAGUGCUACUAGCAACUGCAAUUGCACCAGACCUAU .(((((.((((....(..((.(((((...))))).))..).................((((((.....)))))).....)))).))))).......... (-15.69 = -16.97 + 1.28)


| Location | 5,291,127 – 5,291,221 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -20.86 |
| Energy contribution | -22.58 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
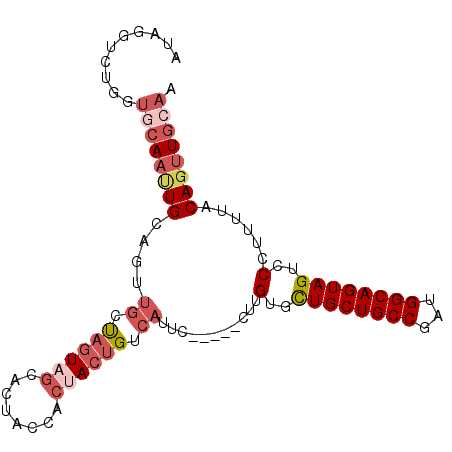
>3L_DroMel_CAF1 5291127 94 - 23771897 AUAGGUUUGCUGCAAUUGCAGUUGCUAGUAGCACUACCACUACUGUCAUUC-----CUUGUGCUGCUGCCGAUGGCAGUAGUUCCUUUUACAGUUGCAA ...(((.((((((....((....))..))))))..)))...(((((.....-----.....(((((((((...))))))))).......)))))..... ( -29.43) >DroSec_CAF1 38546 94 - 1 AUAGGUCUGGUGCAACUGCAGUUGCUAGUAGCACUGCCACUACUGUCAUUC-----CUUGUGCUGCUGCCGAUGGCAGUAGUUCCUUUUACAGUUGCAA ..........((((((((((((.((.....))))))).((((((((((((.-----...(.((....))))))))))))))).........))))))). ( -33.20) >DroSim_CAF1 39373 94 - 1 AUAGGUCUGGUGCAACUGCAGUUGCUAGUAGCACUACCACUACUGUCAUUC-----CUUGCGCUGCUGCCGAUGGCAGUAGUUCCUUUUACAGUUGCAA ..........(((((((((((.((.((((((........)))))).))...-----.))))(((((((((...))))))))).........))))))). ( -31.40) >DroEre_CAF1 38106 81 - 1 -------------AUGUGCAGUUGCUAGUAGCACUACCACUACUGUCAUUC-----CUGGUGCUGCUGCCGAUGGCAGUAGUCCCUUUUACAGUUGCAA -------------...((((..((.((((((........))))))......-----..((..((((((((...))))))))..)).....))..)))). ( -28.20) >DroYak_CAF1 39864 94 - 1 AUAGGUCUUCUGCAAUUGCAGUUGCUAGUAGUACAACCACCACUGUCAUUC-----CUUGUGCUGCUGCCGAUGGCAGUAGUCCCUCUUACAGUUGCAA ..........((((((((....((.((((.((......)).)))).))...-----...(..((((((((...))))))))..)......)))))))). ( -25.40) >DroPer_CAF1 79141 88 - 1 -------CAGUACAAUUGCC----CCAUUCUUGUUGCUGCUACAGCCAUUCUGCUGCUGGUAUUGCUGCCGAUGGCAGUAGCCCCUUUUACAGUUGCAA -------.....((((((..----........((((((((((((((......)))).(((((....))))).))))))))))........))))))... ( -24.07) >consensus AUAGGUCUGGUGCAAUUGCAGUUGCUAGUAGCACUACCACUACUGUCAUUC_____CUUGUGCUGCUGCCGAUGGCAGUAGUCCCUUUUACAGUUGCAA ..........((((((((....((.((((((........)))))).))...........(..((((((((...))))))))..)......)))))))). (-20.86 = -22.58 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:14 2006