| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,270,010 – 5,270,112 |
| Length | 102 |
| Max. P | 0.812440 |

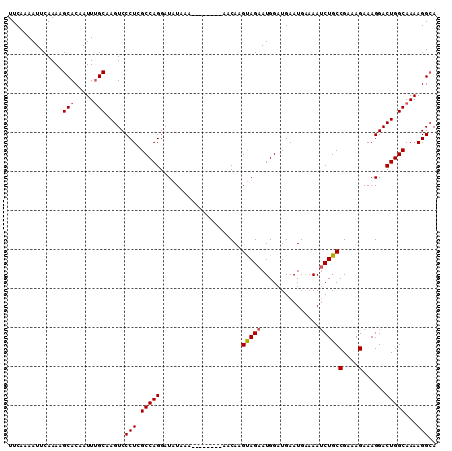
| Location | 5,270,010 – 5,270,112 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
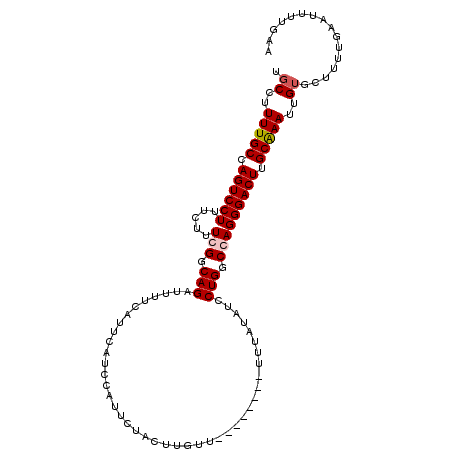
>3L_DroMel_CAF1 5270010 102 + 23771897 UUCCAAAUUCAAAAGCACAAUUUGCAAGUCCCUGGCCAGGAUAUAAA--------AACAAGUAGAAUGGAUGAAUGAAAAUCUGCCGAAAGAAAGGACUGGCAAAAGGCA ..............((....(((((.(((((....(((...(((...--------.....)))...)))...........(((......)))..))))).)))))..)). ( -19.50) >DroSec_CAF1 24773 107 + 1 UUCAAAAUUCAAAAGCACAAUUUGCAAGUCCCUCGCCAGGAUAUAAAAA---AAAAACAAGUAGAAUGGAUGAAUGAAAAACUGCCGAAAGAAAGGACUGGCAAAAGGCA ..............((....(((((.(((((.((((((...(((.....---........)))...))).)))............(....)...))))).)))))..)). ( -19.82) >DroSim_CAF1 24454 110 + 1 UUCAAAAUUCAAAAGCACAAUUUGCAAGUCCCUCGCCAGGAUAUAAAAAAAAAAAAACAAGUAGAAUGGAUGAAUGAAAAUCUGCCGAAAGAAAGGACUGGCAAAAGGCA ..............((....(((((.(((((.((((((...(((................)))...))).))).......(((......)))..))))).)))))..)). ( -19.69) >DroEre_CAF1 24999 101 + 1 UUCAAAAUUCAAAAGCACAAUUCGCCAGUCCCUGGCCAGGAUAUAAA--------AACAAGCAGAAUGGCUGAAUGGAAAUCUGCCGAAAGAAAGGACUGGCAAAAGG-A .......................((((((((................--------.....(((((....((....))...)))))(....)...))))))))......-. ( -24.10) >consensus UUCAAAAUUCAAAAGCACAAUUUGCAAGUCCCUCGCCAGGAUAUAAA________AACAAGUAGAAUGGAUGAAUGAAAAUCUGCCGAAAGAAAGGACUGGCAAAAGGCA ..............(((.....))).....(((.(((((.....................(((((...............)))))(....)......)))))...))).. (-15.87 = -16.18 + 0.31)


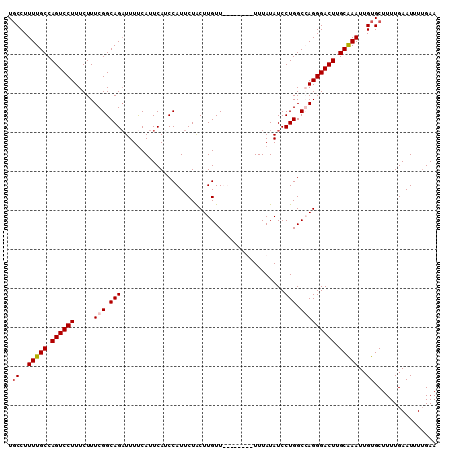
| Location | 5,270,010 – 5,270,112 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -15.74 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5270010 102 - 23771897 UGCCUUUUGCCAGUCCUUUCUUUCGGCAGAUUUUCAUUCAUCCAUUCUACUUGUU--------UUUAUAUCCUGGCCAGGGACUUGCAAAUUGUGCUUUUGAAUUUGGAA .(((.(((((.(((((((......(((.(((.......((...........))..--------.....)))...)))))))))).)))))..).)).............. ( -19.14) >DroSec_CAF1 24773 107 - 1 UGCCUUUUGCCAGUCCUUUCUUUCGGCAGUUUUUCAUUCAUCCAUUCUACUUGUUUUU---UUUUUAUAUCCUGGCGAGGGACUUGCAAAUUGUGCUUUUGAAUUUUGAA .(((.(((((.((((((.....(((.(((..(..........................---.......)..))).))))))))).)))))..).)).............. ( -19.71) >DroSim_CAF1 24454 110 - 1 UGCCUUUUGCCAGUCCUUUCUUUCGGCAGAUUUUCAUUCAUCCAUUCUACUUGUUUUUUUUUUUUUAUAUCCUGGCGAGGGACUUGCAAAUUGUGCUUUUGAAUUUUGAA .(((.(((((.((((((.....(((.(((..........................................))).))))))))).)))))..).)).............. ( -19.95) >DroEre_CAF1 24999 101 - 1 U-CCUUUUGCCAGUCCUUUCUUUCGGCAGAUUUCCAUUCAGCCAUUCUGCUUGUU--------UUUAUAUCCUGGCCAGGGACUGGCGAAUUGUGCUUUUGAAUUUUGAA .-...(((((((((((((......(((.((.......)).))).....(((.(..--------(....)..).))).))))))))))))).................... ( -26.50) >consensus UGCCUUUUGCCAGUCCUUUCUUUCGGCAGAUUUUCAUUCAUCCAUUCUACUUGUU________UUUAUAUCCUGGCCAGGGACUUGCAAAUUGUGCUUUUGAAUUUUGAA .((..(((((.((((((.....(((.(((..........................................))).))))))))).)))))..))................ (-15.74 = -16.30 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:06 2006