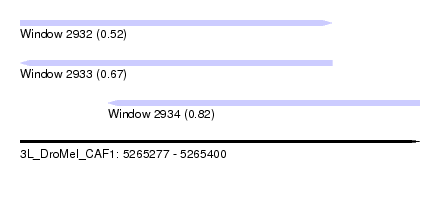
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,265,277 – 5,265,400 |
| Length | 123 |
| Max. P | 0.819543 |

| Location | 5,265,277 – 5,265,373 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -12.76 |
| Consensus MFE | -7.78 |
| Energy contribution | -8.11 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5265277 96 + 23771897 ----------ACAACAAAUAAGGCGAAAACCC-CAAAA----AAUGCGGCAAUAAAAUUGUAUUUAUUACAUUUCUUGAACUUUGCCACAUGAAAAAUUCAAAAGCA--AAAA ----------............((........-.....----.(((.(((((......((((.....))))...........))))).))).............)).--.... ( -11.36) >DroPse_CAF1 55669 100 + 1 AAGUACAAGAACAACA-AUCAG------CCCC-CAAAA----AAUGUGGCAAUAAAAUUGUAUUUAUUACAUUUCUUGAACUUUGCCACAUGAAAAAUUCAAAAGAAACCGA- ................-.....------....-.....----.(((((((((......((((.....))))...........))))))))).....................- ( -13.23) >DroEre_CAF1 20307 99 + 1 ----------ACAACA-AUAUGGCGAAAACCCAAAAAAAAAAAUUGCGGCAAUAAAAUUGUAUUUAUUACAUUUCUUGAACUUUGCCACAUGAAAAAUUCAAAAGCA--CAA- ----------......-...(((.(....))))...........((((((((......((((.....))))...........)))))...(((.....)))...)))--...- ( -11.03) >DroYak_CAF1 20576 95 + 1 ----------ACAACA-AUAUGCCGAAAAACCCCAAAA----AAUGCGGCAAUAAAAUUGUAUUUAUUACAUUUCUUGAACUUUGCCACAUGAAAAAUUCAAAAGCA--CAA- ----------......-...(((...............----.(((.(((((......((((.....))))...........))))).)))((.....))....)))--...- ( -10.33) >DroAna_CAF1 20006 95 + 1 ----------ACAACA-AUAUGGCAAAGCCGACAAAAA----AAUUCGGCAAUAAAAUUGUAUUUAUUACAUUUCUUGAACUUUGCCACAUGAAAAAUUCAAAAGCA--CAA- ----------......-...((((((((((((......----...)))))........((((.....))))..........)))))))...................--...- ( -17.40) >DroPer_CAF1 57976 100 + 1 AAGUACAAGAACAACA-AUCAG------CCCC-CAAAA----AAUGUGGCAAUAAAAUUGUAUUUAUUACAUUUCUUGAACUUUGCCACAUGAAAAAUUCAAAAGAAACCGA- ................-.....------....-.....----.(((((((((......((((.....))))...........))))))))).....................- ( -13.23) >consensus __________ACAACA_AUAAGGCGAAAACCC_CAAAA____AAUGCGGCAAUAAAAUUGUAUUUAUUACAUUUCUUGAACUUUGCCACAUGAAAAAUUCAAAAGCA__CAA_ ...........................................(((.(((((......((((.....))))...........))))).)))...................... ( -7.78 = -8.11 + 0.33)



| Location | 5,265,277 – 5,265,373 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5265277 96 - 23771897 UUUU--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAUU----UUUUG-GGGUUUUCGCCUUAUUUGUUGU---------- .(((--((((..(((.....)))...)))))))..((((((((((((((((......))))))...)))))----)))))-((((....))))..........---------- ( -20.00) >DroPse_CAF1 55669 100 - 1 -UCGGUUUCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCACAUU----UUUUG-GGGG------CUGAU-UGUUGUUCUUGUACUU -(((((..((((((((((((((....)).)))))))))))...((((((((......))))))))......----.....-)..)------)))).-................ ( -21.30) >DroEre_CAF1 20307 99 - 1 -UUG--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAAUUUUUUUUUUUGGGUUUUCGCCAUAU-UGUUGU---------- -...--................(((((((((((..(((((((.((((((((......))))))))..........)))))))..)))).)))))))-......---------- ( -21.40) >DroYak_CAF1 20576 95 - 1 -UUG--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAUU----UUUUGGGGUUUUUCGGCAUAU-UGUUGU---------- -.((--((((...(((((((..(((((((((......((((.((((.....)))).)))).))))))))).----....)))))))...)))))).-......---------- ( -20.40) >DroAna_CAF1 20006 95 - 1 -UUG--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGAAUU----UUUUUGUCGGCUUUGCCAUAU-UGUUGU---------- -...--................((((((((((((.(((((((.((((((((......)))))))).....)----))))))...))))))))))))-......---------- ( -25.00) >DroPer_CAF1 57976 100 - 1 -UCGGUUUCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCACAUU----UUUUG-GGGG------CUGAU-UGUUGUUCUUGUACUU -(((((..((((((((((((((....)).)))))))))))...((((((((......))))))))......----.....-)..)------)))).-................ ( -21.30) >consensus _UUG__UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAUU____UUUUG_GGGGUUUCGCCAUAU_UGUUGU__________ .........(((((((((((((....)).)))))))))))...((((((((......))))))))................................................ (-12.70 = -12.70 + 0.00)


| Location | 5,265,304 – 5,265,400 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5265304 96 - 23771897 --CGU-UGCA-CACACUCG-----UUC--UUUUUUUUUUUUU--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAUU-- --...-(((.-........-----...--.((((((...(((--((((..(((.....)))...)))))))...)))))).((((((((......)))))))).)))..-- ( -17.40) >DroPse_CAF1 55699 93 - 1 C--AGCAAAAGCUGUCUUG-----UUC--UUU-------UCGGUUUCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCACAUU-- (--(((....)))).....-----...--...-------...((((((..((((((((((....)).))))))))))))))((((((((......))))))))......-- ( -18.10) >DroEre_CAF1 20336 89 - 1 --UGUGUGUU----UCUCG-----UUC--UUU-------UUG--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAAUUU --((((.(((----(((.(-----..(--((.-------...--((((..(((.....)))...)))))))..).))))))((((((((......)))))))))))).... ( -18.10) >DroYak_CAF1 20603 89 - 1 --UGUGUGUA----GCUCG-----UUUCGUAU-------UUG--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAUU-- --.(((((..----....(-----(((((((.-------...--)))..(((((((((((....)).))))))))))))))((((((((......))))))))))))).-- ( -17.80) >DroAna_CAF1 20033 94 - 1 GAAACCAGUA----GCCGGUUUUUUUU--UUU-------UUG--UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGAAUU-- ((((((....----...))))))....--..(-------(((--...(((((((((((((....)).)))))))))))...((((((((......))))))))))))..-- ( -16.70) >DroPer_CAF1 58006 93 - 1 C--AGCAAAAGCUGUCUUG-----CUC--UUU-------UCGGUUUCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCACAUU-- (--(((....)))).....-----...--...-------...((((((..((((((((((....)).))))))))))))))((((((((......))))))))......-- ( -18.10) >consensus ___AUCAGUA____UCUCG_____UUC__UUU_______UUG__UGCUUUUGAAUUUUUCAUGUGGCAAAGUUCAAGAAAUGUAAUAAAUACAAUUUUAUUGCCGCAUU__ ...............................................(((((((((((((....)).)))))))))))...((((((((......))))))))........ (-12.70 = -12.70 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:03 2006