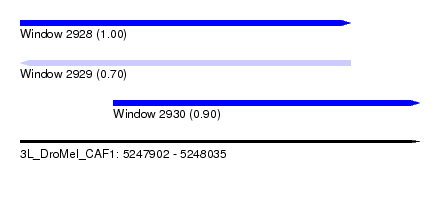
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,247,902 – 5,248,035 |
| Length | 133 |
| Max. P | 0.999580 |

| Location | 5,247,902 – 5,248,012 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.42 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
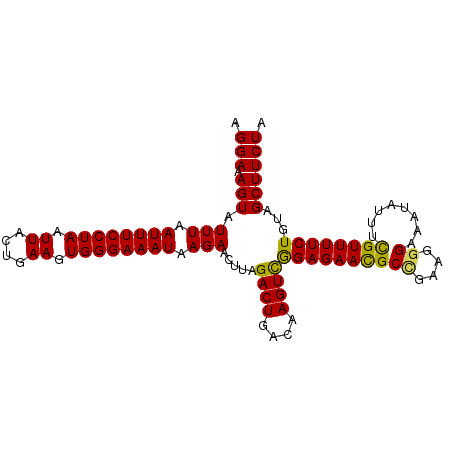
>3L_DroMel_CAF1 5247902 110 + 23771897 AGUGUAAAUGAUAUUUCCUUUUGGACUCUGCAGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUCGGAGAAUGCCGAAGGGAAA .............(((((((((((..(((.(....((((.(((.((((((((.((....)).)))))))).))))))).((((....))))).)))...))))))))))) ( -31.20) >DroSec_CAF1 2594 108 + 1 AGUGUAAAUGAUAUUUCCUUUUGGAC--UGCAGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUCGGAGAACGCCGAAGGGAAA .............(((((((((((.(--(.(....((((.(((.((((((((.((....)).)))))))).))))))).((((....))))).))....))))))))))) ( -28.50) >DroSim_CAF1 704 108 + 1 AGUGUAAAUGAUAUUUCCUUUUGGAC--UGCAGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUUGGAGAACGCCGAAGGGAAA .............(((((((((((..--(((((..((((.(((.((((((((.((....)).)))))))).)))))))...))).)).(((....))).))))))))))) ( -26.50) >DroEre_CAF1 721 107 + 1 AGUGUAAAUGAUAUUUCCUUUUGGAC--AGCAGGAAAGUAUUUAAUUUCCUAAUUACUGAAAUGGGAAAUAAGAACUUAGACUGACAAGUCAGAGAACGCCGAAG-GAAA .((((......((((((((((..(..--...((((((........)))))).....)..))..))))))))....(((.((((....)))).))).)))).....-.... ( -24.50) >DroYak_CAF1 2694 108 + 1 AGUGUAAAUGAUAUUUCCUUUCGGAC--UGCAGGAUAGUAUUUAAUUUCCUAAUUACUGAAAUGGGAAAUAAGAAGUUAGACUGACAAGUCGGAGAACGCUGAAGGGAAA .............(((((((((((((--(.(((..(((..(((.((((((((.((....)).)))))))).)))..)))..)))...))))(.....)..)))))))))) ( -28.50) >consensus AGUGUAAAUGAUAUUUCCUUUUGGAC__UGCAGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUCGGAGAACGCCGAAGGGAAA .............(((((((((((................(((.((((((((.((....)).)))))))).))).(((.((((....)))).)))....))))))))))) (-24.50 = -24.42 + -0.08)

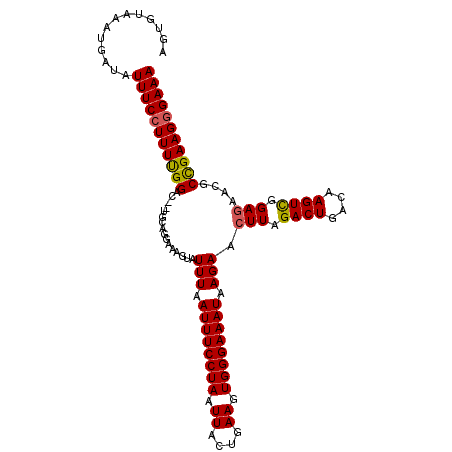

| Location | 5,247,902 – 5,248,012 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -17.22 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5247902 110 - 23771897 UUUCCCUUCGGCAUUCUCCGACUUGUCAGUCUAAGUUCUUAUUUCCCACUUCAGUAAUUAGGAAAUUAAAUACUUUCCUGCAGAGUCCAAAAGGAAAUAUCAUUUACACU (((((....((......))(((((..(((...((((....((((((..............)))))).....))))..)))..))))).....)))))............. ( -18.54) >DroSec_CAF1 2594 108 - 1 UUUCCCUUCGGCGUUCUCCGACUUGUCAGUCUAAGUUCUUAUUUCCCACUUCAGUAAUUAGGAAAUUAAAUACUUUCCUGCA--GUCCAAAAGGAAAUAUCAUUUACACU .......((((......))))......(((.(((((...(((((((.((....))...(((((((........)))))))..--........)))))))..))))).))) ( -17.60) >DroSim_CAF1 704 108 - 1 UUUCCCUUCGGCGUUCUCCAACUUGUCAGUCUAAGUUCUUAUUUCCCACUUCAGUAAUUAGGAAAUUAAAUACUUUCCUGCA--GUCCAAAAGGAAAUAUCAUUUACACU .........((((..........))))(((.(((((...(((((((.((....))...(((((((........)))))))..--........)))))))..))))).))) ( -16.20) >DroEre_CAF1 721 107 - 1 UUUC-CUUCGGCGUUCUCUGACUUGUCAGUCUAAGUUCUUAUUUCCCAUUUCAGUAAUUAGGAAAUUAAAUACUUUCCUGCU--GUCCAAAAGGAAAUAUCAUUUACACU ((((-((((((((..((..((((....))))..))..).....................((((((........)))))))))--).....)))))))............. ( -19.40) >DroYak_CAF1 2694 108 - 1 UUUCCCUUCAGCGUUCUCCGACUUGUCAGUCUAACUUCUUAUUUCCCAUUUCAGUAAUUAGGAAAUUAAAUACUAUCCUGCA--GUCCGAAAGGAAAUAUCAUUUACACU (((((.(((...(.....)((((.(.(((..((.......((((((..............)))))).......))..)))))--))).))).)))))............. ( -14.38) >consensus UUUCCCUUCGGCGUUCUCCGACUUGUCAGUCUAAGUUCUUAUUUCCCACUUCAGUAAUUAGGAAAUUAAAUACUUUCCUGCA__GUCCAAAAGGAAAUAUCAUUUACACU .........((((..........))))(((.(((((...(((((((............(((((((........)))))))............)))))))..))))).))) (-14.79 = -15.23 + 0.44)

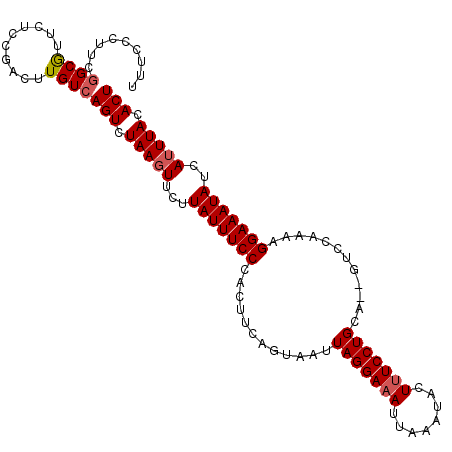
| Location | 5,247,933 – 5,248,035 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 95.59 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.14 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5247933 102 + 23771897 AGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUCGGAGAAUGCCGAAGGGAAAUAUUUCGUUUUCUGUAGCUUCUA ((((((........)))))).......(((((.(((((...((((((.((((....)))).)))....((....))......)))..)))))...))))).. ( -24.90) >DroSec_CAF1 2623 102 + 1 AGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUCGGAGAACGCCGAAGGGAAAUAUUUCGUUUUCUGUAGCUUCUA ((((((........)))))).......(((((.(((((...((((((.((((....)))).)))....((....))......)))..)))))...))))).. ( -24.90) >DroSim_CAF1 733 102 + 1 AGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUUGGAGAACGCCGAAGGGAAAUAUUUCGUUUUCUGUAGCUUCUA ((((((........)))))).......(((((.(((((...((((((.((((....)))).)))....((....))......)))..)))))...))))).. ( -23.20) >DroEre_CAF1 750 101 + 1 AGGAAAGUAUUUAAUUUCCUAAUUACUGAAAUGGGAAAUAAGAACUUAGACUGACAAGUCAGAGAACGCCGAAG-GAAAUAUUUUGUUUUCUGAAGCUUCUA ....((((.(((.((((((((.((....)).)))))))).))))))).((((....))))..((((.((...((-(((((.....)))))))...)))))). ( -22.40) >DroYak_CAF1 2723 102 + 1 AGGAUAGUAUUUAAUUUCCUAAUUACUGAAAUGGGAAAUAAGAAGUUAGACUGACAAGUCGGAGAACGCUGAAGGGAAAUAUUUCGUUUUCUGCAGCUUCUA .............((((((((.((....)).)))))))).(((((((.((((....))))((((((((..(((........)))))))))))..))))))). ( -24.60) >consensus AGGAAAGUAUUUAAUUUCCUAAUUACUGAAGUGGGAAAUAAGAACUUAGACUGACAAGUCGGAGAACGCCGAAGGGAAAUAUUUCGUUUUCUGUAGCUUCUA .(((.(((.(((.((((((((.((....)).)))))))).))).....((((....))))((((((((((....))........))))))))...)))))). (-19.70 = -19.14 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:59 2006