
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,242,496 – 5,242,588 |
| Length | 92 |
| Max. P | 0.781338 |

| Location | 5,242,496 – 5,242,588 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
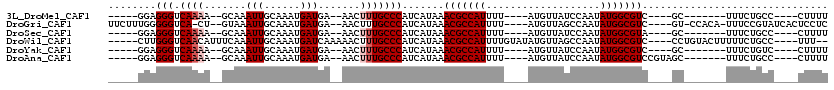
>3L_DroMel_CAF1 5242496 92 + 23771897 -----GGAGGGUCAAAA--GCAAAUUGCAAAUGAUGA--AACUUUGCCCAUCAUAAACGCCAUUUU----AUGUUAUCCAAUAUGGCGUC----GC-------UUUCUGCC----CUUUU -----(((((((..(((--((.........((((((.--.........))))))..(((((((...----............))))))).----))-------)))..)))----)))). ( -27.86) >DroGri_CAF1 130769 105 + 1 UUCUUUGGGGGUCA-CU--GUAAAUUGCAAAUGAUGA--AACUUUGCCCAUCAUAAACGCCAUUUU----AUGUUAGCCAAUAUGGCGUC----GU-CCACA-UUUCCGUAUCACUCCUC ......((((((..-..--...........((((((.--.........))))))..(((((((...----............))))))).----..-.....-..........)))))). ( -18.86) >DroSec_CAF1 106806 92 + 1 -----GGAGGGUCAAAA--GCAAAUUGCAAAUGAUGA--AACUUUGCCCAUCAUAAACGCCAUUUU----AUGUUAUCCAAUAUGGCGUA----GC-------UUUCUGCC----CUUUU -----(((((((..(((--((.........((((((.--.........))))))..(((((((...----............))))))).----))-------)))..)))----)))). ( -28.36) >DroWil_CAF1 144426 105 + 1 -----CUUGGGUCAACAUUUCAAAUUGCAAAUGAUCAAAAACUUUGCCCAUCAUAAACGCCAUUUUGUAUAUGUUAGCCAAUAUGGCGUC----CCUGUACUUUUUCUGCC----UUU-- -----...(((.((((((..((((..((..((((((((.....)))...)))))....))...))))...))))).(((.....)))).)----))...............----...-- ( -17.90) >DroYak_CAF1 109254 92 + 1 -----GGAGGGUCAAAA--GCAAAUUGCAAAUGAUGA--AACUUUGCCCAUCAUAAACGCCAUUUU----AUGUUAUCCAAUAUGGCGUC----GC-------UUUCUGUC----CUUUU -----.(((((.(((((--((.........((((((.--.........))))))..(((((((...----............))))))).----))-------))).)).)----)))). ( -24.46) >DroAna_CAF1 104318 96 + 1 -----GGAGGGUCAAAA--GCAAAUUGCAAAUGAUGA--AACUUUGCCCAUCAUAAACGCCAUUUU----AUGUUAUCCAAUAUGGCGUCCGUAGC-------UUUCUGCC----CUUUU -----(((((((..(((--((.........((((((.--.........))))))..(((((((...----............))))))).....))-------)))..)))----)))). ( -27.76) >consensus _____GGAGGGUCAAAA__GCAAAUUGCAAAUGAUGA__AACUUUGCCCAUCAUAAACGCCAUUUU____AUGUUAUCCAAUAUGGCGUC____GC_______UUUCUGCC____CUUUU ........(((.((((.......(((......))).......))))))).......(((((((...................)))))))............................... (-13.38 = -13.88 + 0.50)

| Location | 5,242,496 – 5,242,588 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -13.61 |
| Energy contribution | -14.14 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
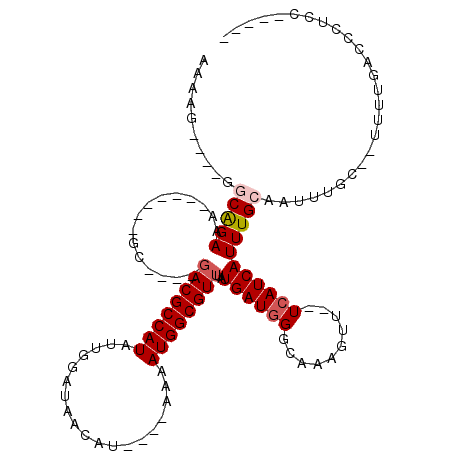
>3L_DroMel_CAF1 5242496 92 - 23771897 AAAAG----GGCAGAAA-------GC----GACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAGUU--UCAUCAUUUGCAAUUUGC--UUUUGACCCUCC----- ...((----((..((((-------((----((.(((((.(((.......)----)).)))))((..((((((((......)--)))))))..))...))))--))))..))))..----- ( -30.80) >DroGri_CAF1 130769 105 - 1 GAGGAGUGAUACGGAAA-UGUGG-AC----GACGCCAUAUUGGCUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAGUU--UCAUCAUUUGCAAUUUAC--AG-UGACCCCCAAAGAA (.((.((.((...(((.-((..(-(.----((((((((.(((.......)----)).))))))))..(((((((......)--))))))))..)).)))..--.)-).)).)))...... ( -25.20) >DroSec_CAF1 106806 92 - 1 AAAAG----GGCAGAAA-------GC----UACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAGUU--UCAUCAUUUGCAAUUUGC--UUUUGACCCUCC----- ...((----((..((((-------((----...(((((.(((.......)----)).)))))((..((((((((......)--)))))))..)).....))--))))..))))..----- ( -29.10) >DroWil_CAF1 144426 105 - 1 --AAA----GGCAGAAAAAGUACAGG----GACGCCAUAUUGGCUAACAUAUACAAAAUGGCGUUUAUGAUGGGCAAAGUUUUUGAUCAUUUGCAAUUUGAAAUGUUGACCCAAG----- --...----.............((..----((((((((.(((...........))).))))))))..)).(((((((.(((((.(((........))).))))).))).))))..----- ( -26.10) >DroYak_CAF1 109254 92 - 1 AAAAG----GACAGAAA-------GC----GACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAGUU--UCAUCAUUUGCAAUUUGC--UUUUGACCCUCC----- ...((----(...((((-------((----((.(((((.(((.......)----)).)))))((..((((((((......)--)))))))..))...))))--))))...)))..----- ( -26.70) >DroAna_CAF1 104318 96 - 1 AAAAG----GGCAGAAA-------GCUACGGACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAGUU--UCAUCAUUUGCAAUUUGC--UUUUGACCCUCC----- ...((----((..((((-------((...(((((((((.(((.......)----)).)))))))))((((((((......)--))))))).........))--))))..))))..----- ( -30.40) >consensus AAAAG____GGCAGAAA_______GC____GACGCCAUAUUGGAUAACAU____AAAAUGGCGUUUAUGAUGGGCAAAGUU__UCAUCAUUUGCAAUUUGC__UUUUGACCCUCC_____ ..........(((((...............((((((((...................))))))))..((((((..........))))))))))).......................... (-13.61 = -14.14 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:54 2006