
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
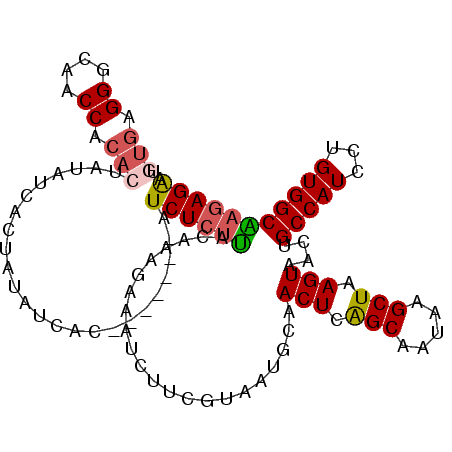
| Location | 5,238,491 – 5,238,596 |
| Length | 105 |
| Max. P | 0.973249 |

| Location | 5,238,491 – 5,238,596 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
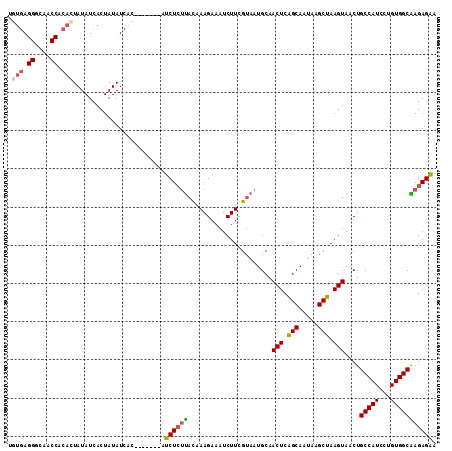
>3L_DroMel_CAF1 5238491 105 + 23771897 UGUGAGGGCAACCACACUAUAUCACUAUAUCAC-------AUCUCUUACAAAGAAAUCUUCGUAAUGCAACUCAGCAAUAAGCUAAGUAACUGCCAUCCUGUGGCAAGAGAA (((((((....))....((((....))))))))-------)((((((((.(((....))).))).....(((.(((.....))).)))...((((((...))))))))))). ( -25.90) >DroSec_CAF1 102902 105 + 1 UGUGAGGGCAACCACACUAUAUCACUAUAUCAC-------AUCUCUUUCAAAGAAAUCUUCGUAAUGCAACUCAGCAAUAAGCUAAGUAACUGCCAUCCUGUGGCAAGAGAA (((((((....))....((((....))))))))-------)((((((......................(((.(((.....))).)))....(((((...))))))))))). ( -24.00) >DroSim_CAF1 98970 105 + 1 UGUGAGGGCAACCACACUAUAUCACUAUAUCAC-------AUCUCUUACAAAGAAAUCUUCGUAAUGCAACUCAGCAAUAAGCUAAGUAACUGCCAUCCUGUGGCAAGAGAA (((((((....))....((((....))))))))-------)((((((((.(((....))).))).....(((.(((.....))).)))...((((((...))))))))))). ( -25.90) >DroEre_CAF1 105888 112 + 1 UGUGAGGGCAACCACACUAUAUCACUAUAUCUCUCUAACAAUCUCUUACAAAGAAAUCUUCGUAAUGCAACUCAGCAAUAAGCUAAGUAACUGCCAUCCUGUGGCGAGAGGA .(((.((....)).)))............((((((..........((((.(((....))).))))....(((.(((.....))).)))....(((((...))))))))))). ( -28.50) >DroYak_CAF1 105409 101 + 1 ----AGGGCAACCACACUAUAUCACUAUAUCAC-------AUCUCUUACAAAGAAAUCUUCGUAAUGCAACUCAGCAAUAAGCUAAGUAACUGCCAUCCUGUGGCAAGAGAA ----.((....))....................-------.((((((((.(((....))).))).....(((.(((.....))).)))...((((((...))))))))))). ( -22.40) >DroAna_CAF1 100309 104 + 1 UAUGAGGGCAACCGCACUAUAUCACUAUAUCAU-------AUCUAGGAUAAAGAAAUCUUUGUAAUGCAACUCGGCAAUAAGCUAAGUAACUGCCAUCCUGUGGCCAGAGA- ..((.((....)).))((.((((.(((......-------...))))))).))...((((((.......(((..((.....))..)))....(((((...)))))))))))- ( -23.30) >consensus UGUGAGGGCAACCACACUAUAUCACUAUAUCAC_______AUCUCUUACAAAGAAAUCUUCGUAAUGCAACUCAGCAAUAAGCUAAGUAACUGCCAUCCUGUGGCAAGAGAA .(((.((....)).)))........................((((((......................(((.(((.....))).)))....(((((...))))))))))). (-21.07 = -21.40 + 0.33)

| Location | 5,238,491 – 5,238,596 |
|---|---|
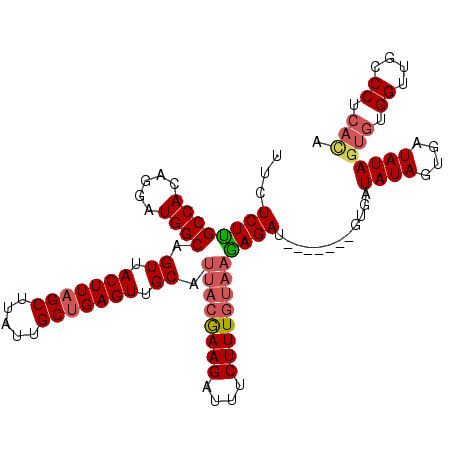
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -27.02 |
| Energy contribution | -27.72 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
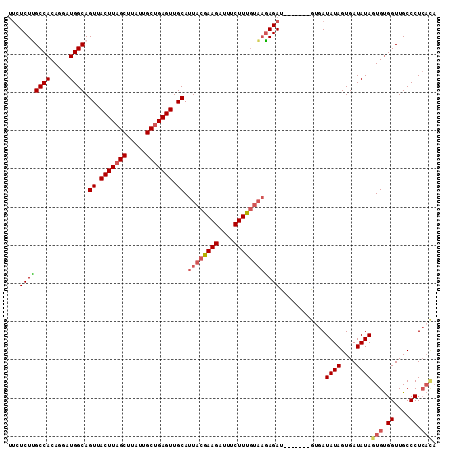
>3L_DroMel_CAF1 5238491 105 - 23771897 UUCUCUUGCCACAGGAUGGCAGUUACUUAGCUUAUUGCUGAGUUGCAUUACGAAGAUUUCUUUGUAAGAGAU-------GUGAUAUAGUGAUAUAGUGUGGUUGCCCUCACA .((((.(((((.....)))))((.(((((((.....))))))).)).((((((((....)))))))))))).-------....((((....))))(((.((....)).))). ( -34.40) >DroSec_CAF1 102902 105 - 1 UUCUCUUGCCACAGGAUGGCAGUUACUUAGCUUAUUGCUGAGUUGCAUUACGAAGAUUUCUUUGAAAGAGAU-------GUGAUAUAGUGAUAUAGUGUGGUUGCCCUCACA .((((((((((.....)))).((.(((((((.....))))))).))....(((((....))))).)))))).-------....((((....))))(((.((....)).))). ( -32.20) >DroSim_CAF1 98970 105 - 1 UUCUCUUGCCACAGGAUGGCAGUUACUUAGCUUAUUGCUGAGUUGCAUUACGAAGAUUUCUUUGUAAGAGAU-------GUGAUAUAGUGAUAUAGUGUGGUUGCCCUCACA .((((.(((((.....)))))((.(((((((.....))))))).)).((((((((....)))))))))))).-------....((((....))))(((.((....)).))). ( -34.40) >DroEre_CAF1 105888 112 - 1 UCCUCUCGCCACAGGAUGGCAGUUACUUAGCUUAUUGCUGAGUUGCAUUACGAAGAUUUCUUUGUAAGAGAUUGUUAGAGAGAUAUAGUGAUAUAGUGUGGUUGCCCUCACA ..(((((((((.....)))).((.(((((((.....))))))).)).((((((((....))))))))..........))))).((((....))))(((.((....)).))). ( -37.50) >DroYak_CAF1 105409 101 - 1 UUCUCUUGCCACAGGAUGGCAGUUACUUAGCUUAUUGCUGAGUUGCAUUACGAAGAUUUCUUUGUAAGAGAU-------GUGAUAUAGUGAUAUAGUGUGGUUGCCCU---- ...(((((...))))).(((((..((...((((((..(((.((..((((((((((....))))))....)))-------)..)).)))..))).)))))..)))))..---- ( -30.60) >DroAna_CAF1 100309 104 - 1 -UCUCUGGCCACAGGAUGGCAGUUACUUAGCUUAUUGCCGAGUUGCAUUACAAAGAUUUCUUUAUCCUAGAU-------AUGAUAUAGUGAUAUAGUGCGGUUGCCCUCAUA -...(((....)))((.(((((..(((..((.....))..)))(((((((.((((....))))((((((...-------......))).))).))))))).))))).))... ( -24.10) >consensus UUCUCUUGCCACAGGAUGGCAGUUACUUAGCUUAUUGCUGAGUUGCAUUACGAAGAUUUCUUUGUAAGAGAU_______GUGAUAUAGUGAUAUAGUGUGGUUGCCCUCACA ...((((((((.....)))).((.(((((((.....))))))).)).((((((((....))))))))))))............((((....))))(((.((....)).))). (-27.02 = -27.72 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:49 2006