
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
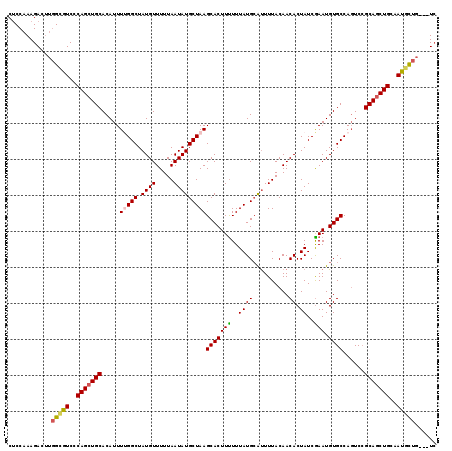
| Location | 5,237,688 – 5,237,804 |
| Length | 116 |
| Max. P | 0.907936 |

| Location | 5,237,688 – 5,237,804 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5237688 116 + 23771897 CUCCAAAGACUUGGCGUCCCAGCUGCACAUUUUGGCUAUGUUUUUAAUAUGCUAAGCACUUUUUUAUGCAUUUUACAACACUAUCGAAUGUGCCAGUCCGCAGCUGCAAUGCUG---UC .......(((..(((((..(((((((.....(((((...(((..(((.((((((((......)))).)))).))).)))((........)))))))...)))))))..))))))---)) ( -29.70) >DroSec_CAF1 102084 116 + 1 CUCCAAAGAAUUGGCGUCCCAGCUGCACAUUUUGGCUAUGUUUUUAAUAUGCUAAGCACUUUUUUAUGAAUUUUACAACACUAUCGAAUGUGCCAGUCCGCAGCUGCAAUGCUG---UC ............(((((..(((((((.....(((((.((((.....)))))))))((((..(((.(((.............))).))).))))......)))))))..))))).---.. ( -27.32) >DroEre_CAF1 105100 119 + 1 CUCCAAAGACUUGGUGUCCCAGCUGCACAUUUCGGCUAUGUUUUUAAUAUGCUAAGCACUUCUUUAUGCGUUUUACAACACUAUCGAAUGUGCCAGUCCGCAGCUGCAAUGCUGUCGUC .......(((..(((((..(((((((.......(((.((((.....)))))))..(((((((..((.(.(((....))).)))..))).))))......)))))))..))))))))... ( -29.50) >DroYak_CAF1 104652 116 + 1 CUCCAAAGACUUGGCGUCCCAGCUGCACAUUUUGGCUAUGUUUUUAAUAUGCUAAGCACUUUUUUAUGCGCUUUACAACACUAUCGAAUGUGCCAGUCCGCAGCUGCAAUGCUG---UC .......(((..(((((..(((((((.....(((((.((((((......((..((((.(........).))))..))........)))))))))))...)))))))..))))))---)) ( -31.74) >DroAna_CAF1 99695 116 + 1 CCCCAAAGACAACGCGUCGCAGCUGCACAUUUCGGCUAUGUUUUUAAUAUGCUAAGCACUUUUUUAUGCAUUUUACAACACUAUCAAAUGUGCCACUCAGCAACUGCAACAUCC---UG .......(((.....)))((((((((((((((.((...((((..(((.((((((((......)))).)))).))).))))...)))))))))).....)))...))).......---.. ( -21.00) >consensus CUCCAAAGACUUGGCGUCCCAGCUGCACAUUUUGGCUAUGUUUUUAAUAUGCUAAGCACUUUUUUAUGCAUUUUACAACACUAUCGAAUGUGCCAGUCCGCAGCUGCAAUGCUG___UC ............(((((..(((((((.....(((((.((((.....)))))))))(((((((..((((..........)).))..))).))))......)))))))..)))))...... (-23.22 = -23.46 + 0.24)


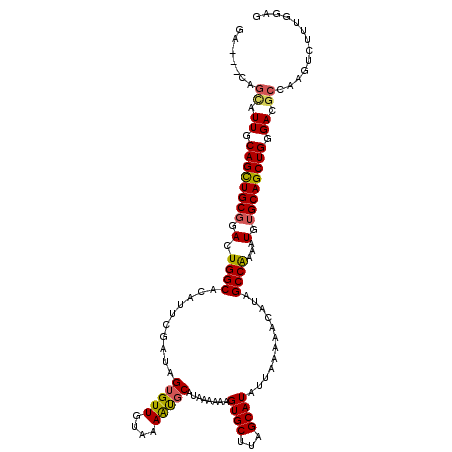
| Location | 5,237,688 – 5,237,804 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -25.84 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5237688 116 - 23771897 GA---CAGCAUUGCAGCUGCGGACUGGCACAUUCGAUAGUGUUGUAAAAUGCAUAAAAAAGUGCUUAGCAUAUUAAAAACAUAGCCAAAAUGUGCAGCUGGGACGCCAAGUCUUUGGAG ((---(.((.((.((((((((.(.((((((((......))))(((.....((((......))))...))).............))))...).)))))))).)).))...)))....... ( -31.10) >DroSec_CAF1 102084 116 - 1 GA---CAGCAUUGCAGCUGCGGACUGGCACAUUCGAUAGUGUUGUAAAAUUCAUAAAAAAGUGCUUAGCAUAUUAAAAACAUAGCCAAAAUGUGCAGCUGGGACGCCAAUUCUUUGGAG ..---..((.((.((((((((.(.((((((((......))))..................((((...))))............))))...).)))))))).)).))(((....)))... ( -27.20) >DroEre_CAF1 105100 119 - 1 GACGACAGCAUUGCAGCUGCGGACUGGCACAUUCGAUAGUGUUGUAAAACGCAUAAAGAAGUGCUUAGCAUAUUAAAAACAUAGCCGAAAUGUGCAGCUGGGACACCAAGUCUUUGGAG (((.......((.(((((((.(...(((((.(((..(((((((....))))).))..))))))))...).........((((.......))))))))))).))......)))....... ( -31.62) >DroYak_CAF1 104652 116 - 1 GA---CAGCAUUGCAGCUGCGGACUGGCACAUUCGAUAGUGUUGUAAAGCGCAUAAAAAAGUGCUUAGCAUAUUAAAAACAUAGCCAAAAUGUGCAGCUGGGACGCCAAGUCUUUGGAG ((---(.((.((.((((((((.(.((((((((......))))(((.((((((........)))))).))).............))))...).)))))))).)).))...)))....... ( -36.90) >DroAna_CAF1 99695 116 - 1 CA---GGAUGUUGCAGUUGCUGAGUGGCACAUUUGAUAGUGUUGUAAAAUGCAUAAAAAAGUGCUUAGCAUAUUAAAAACAUAGCCGAAAUGUGCAGCUGCGACGCGUUGUCUUUGGGG ((---(..((((((((((((....((((...(((((((.(((((......((((......)))).))))))))))))......))))......))))))))))))..)))......... ( -33.10) >consensus GA___CAGCAUUGCAGCUGCGGACUGGCACAUUCGAUAGUGUUGUAAAAUGCAUAAAAAAGUGCUUAGCAUAUUAAAAACAUAGCCAAAAUGUGCAGCUGGGACGCCAAGUCUUUGGAG .......((.((.((((((((.(.((((..........(((((....)))))........((((...))))............))))...).)))))))).)).))............. (-25.84 = -25.72 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:45 2006