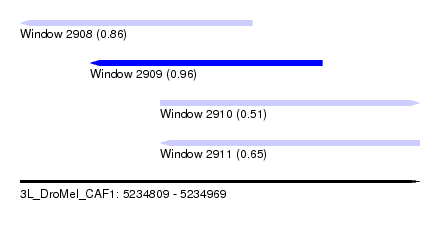
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,234,809 – 5,234,969 |
| Length | 160 |
| Max. P | 0.956108 |

| Location | 5,234,809 – 5,234,902 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 99.28 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -30.89 |
| Energy contribution | -30.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5234809 93 - 23771897 CUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAGAGCCAGGCAACUCUGGCGAGUGAAAAAUCCAAAGCCGG ....((((...((((((((...((((((((..........))))))))..))))))))..))))...(((((...((.......))..))))) ( -30.70) >DroSec_CAF1 99142 93 - 1 CUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAGAGCCAGGCAACUCUGGCGAGUGAAAAAUCCAAAGCCGG ....((((...((((((((...((((((((..........))))))))..))))))))..))))...(((((...((.......))..))))) ( -30.70) >DroEre_CAF1 102153 93 - 1 CUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAAAGCCAGGCAACUCUGGCGAGUGAAAAAUCCAAAGCCGG ....((((...((((((((...((((((((..........))))))))..))))))))..))))...(((((...((.......))..))))) ( -30.60) >consensus CUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAGAGCCAGGCAACUCUGGCGAGUGAAAAAUCCAAAGCCGG ....((((...((((((((...((((((((..........))))))))..))))))))..))))...(((((...((.......))..))))) (-30.89 = -30.67 + -0.22)


| Location | 5,234,837 – 5,234,930 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -23.69 |
| Energy contribution | -23.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5234837 93 - 23771897 CGCAUUUUUUCAGAUUUUCCUGCUUUUCCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAGAGCCAGGCAA .(((......(((......))).......)))((((...((((((((...((((((((..........))))))))..))))))))..)))). ( -26.82) >DroSec_CAF1 99170 84 - 1 CCACUUUUUCCAGAUUUU---------CCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAGAGCCAGGCAA ..........(((.....---------.))).((((...((((((((...((((((((..........))))))))..))))))))..)))). ( -24.50) >DroEre_CAF1 102181 93 - 1 CUUAUUUUUCCAGAUUUUCCUGCUUUUGCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAAAGCCAGGCAA ..........(((......)))..........((((...((((((((...((((((((..........))))))))..))))))))..)))). ( -24.40) >consensus CCAAUUUUUCCAGAUUUUCCUGCUUUUCCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAUGAUGCUGUGUUUAGGAAGAGCCAGGCAA ................................((((...((((((((...((((((((..........))))))))..))))))))..)))). (-23.69 = -23.47 + -0.22)


| Location | 5,234,865 – 5,234,969 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5234865 104 + 23771897 AUCACAAGCUGCAUUAGCAGAAAAGCAGUAGCAGCAGGAAAAGCAGGAAAAUCUGAAAAAAUGCGGGGUGUGGGAAUGUGGAAAUGCAGUACAGAGCUGAACAG .(((((..(((((((..((((...((....)).((.......)).......))))....)))))))..))))).............((((.....))))..... ( -24.00) >DroSec_CAF1 99198 95 + 1 AUCACAAGCUGCAUUAGCAGAAAAGCAGUAGCAGCAGG---------AAAAUCUGGAAAAAGUGGGGGUGUGGGGAUGGGAAAAUGCAGUACAGAGCUGAACAG .(((((.((((((((.((......))))).)))))...---------....(((.(......).))).))))).............((((.....))))..... ( -17.80) >DroEre_CAF1 102209 104 + 1 AUCACAAGCUGCAUUAGCAGAAAAGCAGUAGCAGCAGCAAAAGCAGGAAAAUCUGGAAAAAUAAGGGGUGUGGCGAUGGGCAAAUGCAGUACAGAGCUGCACAG .((.((.((((((((.((......))))).))))).((....)).........))))..........(((..((..((.((....))....))..))..))).. ( -27.20) >consensus AUCACAAGCUGCAUUAGCAGAAAAGCAGUAGCAGCAGGAAAAGCAGGAAAAUCUGGAAAAAUAAGGGGUGUGGGGAUGGGAAAAUGCAGUACAGAGCUGAACAG .(((((.((((((((.((......))))).)))))................(((..........))).))))).............((((.....))))..... (-16.43 = -16.43 + 0.00)


| Location | 5,234,865 – 5,234,969 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -18.07 |
| Consensus MFE | -14.03 |
| Energy contribution | -15.03 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5234865 104 - 23771897 CUGUUCAGCUCUGUACUGCAUUUCCACAUUCCCACACCCCGCAUUUUUUCAGAUUUUCCUGCUUUUCCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAU .......((........)).....((((.....................(((......)))........(((((....((......))....))))).)))).. ( -17.30) >DroSec_CAF1 99198 95 - 1 CUGUUCAGCUCUGUACUGCAUUUUCCCAUCCCCACACCCCCACUUUUUCCAGAUUUU---------CCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAU .......((........)).............((((..............((.....---------.))(((((....((......))....))))).)))).. ( -15.10) >DroEre_CAF1 102209 104 - 1 CUGUGCAGCUCUGUACUGCAUUUGCCCAUCGCCACACCCCUUAUUUUUCCAGAUUUUCCUGCUUUUGCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAU ..((((((.......))))))...........((((.............(((......)))........(((((....((......))....))))).)))).. ( -21.80) >consensus CUGUUCAGCUCUGUACUGCAUUUCCCCAUCCCCACACCCCCAAUUUUUCCAGAUUUUCCUGCUUUUCCUGCUGCUACUGCUUUUCUGCUAAUGCAGCUUGUGAU .......((........)).............((((.............(((......)))........(((((....((......))....))))).)))).. (-14.03 = -15.03 + 1.00)


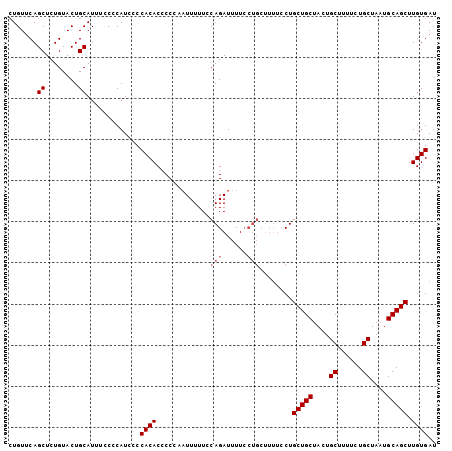
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:41 2006