| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,233,300 – 5,233,402 |
| Length | 102 |
| Max. P | 0.999196 |

| Location | 5,233,300 – 5,233,402 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.85 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
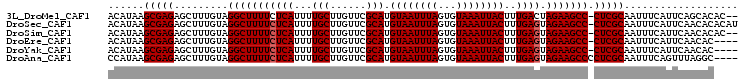
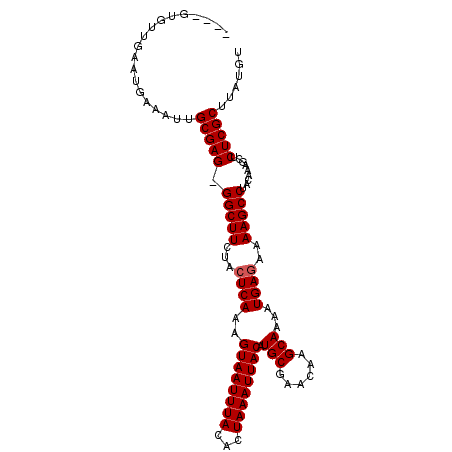
>3L_DroMel_CAF1 5233300 102 + 23771897 ACAUAAGCGAGAGCUUUGUAGGCUUUUCUCAUUUUGCUUGUUCGCAUGUAAUUUAGUGUAAAUUACUUUGACUAGAAGCC-CUCGCAAUUUCAUUCAGCACAC-- ......(((((.(((((.(((.............(((......))).((((((((...)))))))).....)))))))).-))))).................-- ( -22.40) >DroSec_CAF1 97620 104 + 1 ACAUAAGCGAGAGCUUUGUAGGCUUUUCUCAUUUUGCUUGUUCGCAUGUAAUUUAGUGUAAAUUACUUUGAGUAGAAGCC-CUCGCAAUUUCAUUCAACACACAU (((.((((....))))))).(((((((((((...(((......))).((((((((...))))))))..)))).)))))))-........................ ( -25.50) >DroSim_CAF1 94879 102 + 1 ACAUAAGCGAGAGCUUUGUAGGCUUUUCUCAUUUUGCUUGUUCGCAUGUAAUUUAGUGUAAAUUACUUUGAGUAGAAGCC-CUCGCAAUUUCAUUCAACACAC-- (((.((((....))))))).(((((((((((...(((......))).((((((((...))))))))..)))).)))))))-......................-- ( -25.50) >DroEre_CAF1 100644 100 + 1 ACAUAAGCGAGAGCUUUGUAGGCUUUUCUCAUUUUGCUUGUUCGCAUGUAAUUUAGUGUAAAUUACUUUGAGUAGAAGCC-CUCGCAAUUUCAUUCAACAC---- (((.((((....))))))).(((((((((((...(((......))).((((((((...))))))))..)))).)))))))-....................---- ( -25.50) >DroYak_CAF1 100154 100 + 1 ACAUAAGCGAGAGCUUUGUAGGCUUUUCUCAUUUUGCUUGUUCGCAUGUAAUUUAGUGUAAAUUACUUUGAGUAGAAGCC-CUCGCAAUUUCAUUCAACAC---- (((.((((....))))))).(((((((((((...(((......))).((((((((...))))))))..)))).)))))))-....................---- ( -25.50) >DroAna_CAF1 95771 101 + 1 CCAUAAGCGAGAGCUUUGUAGGCUUUUCUCAUUUUGCUUGUUCGCAUGUAAUUUAGUGUAAAUUACUUUGAGUAGAAGCCCCUCGCAAUUUCAGUUUAGGC---- ((.(((((..(((..((((.(((((((((((...(((......))).((((((((...))))))))..)))).)))))))....)))).))).))))))).---- ( -25.90) >consensus ACAUAAGCGAGAGCUUUGUAGGCUUUUCUCAUUUUGCUUGUUCGCAUGUAAUUUAGUGUAAAUUACUUUGAGUAGAAGCC_CUCGCAAUUUCAUUCAACAC____ ......(((((.........(((((((((((...(((......))).((((((((...))))))))..)))).))))))).)))))................... (-23.68 = -23.85 + 0.17)
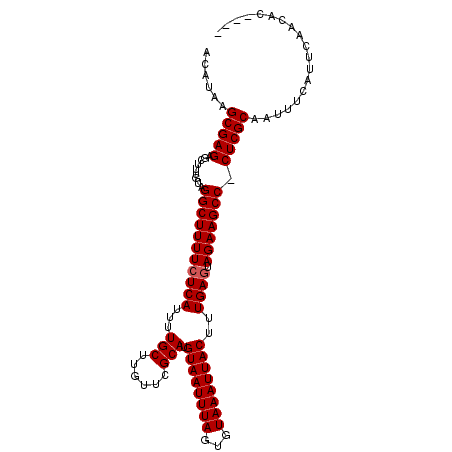
| Location | 5,233,300 – 5,233,402 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5233300 102 - 23771897 --GUGUGCUGAAUGAAAUUGCGAG-GGCUUCUAGUCAAAGUAAUUUACACUAAAUUACAUGCGAACAAGCAAAAUGAGAAAAGCCUACAAAGCUCUCGCUUAUGU --.................(((((-(((((.(((.(...((((((((...)))))))).(((......)))...........).)))..))))))))))...... ( -25.30) >DroSec_CAF1 97620 104 - 1 AUGUGUGUUGAAUGAAAUUGCGAG-GGCUUCUACUCAAAGUAAUUUACACUAAAUUACAUGCGAACAAGCAAAAUGAGAAAAGCCUACAAAGCUCUCGCUUAUGU ...................(((((-(((((...((((..((((((((...)))))))).(((......)))...))))....(....).))))))))))...... ( -25.60) >DroSim_CAF1 94879 102 - 1 --GUGUGUUGAAUGAAAUUGCGAG-GGCUUCUACUCAAAGUAAUUUACACUAAAUUACAUGCGAACAAGCAAAAUGAGAAAAGCCUACAAAGCUCUCGCUUAUGU --.................(((((-(((((...((((..((((((((...)))))))).(((......)))...))))....(....).))))))))))...... ( -25.60) >DroEre_CAF1 100644 100 - 1 ----GUGUUGAAUGAAAUUGCGAG-GGCUUCUACUCAAAGUAAUUUACACUAAAUUACAUGCGAACAAGCAAAAUGAGAAAAGCCUACAAAGCUCUCGCUUAUGU ----...............(((((-(((((...((((..((((((((...)))))))).(((......)))...))))....(....).))))))))))...... ( -25.60) >DroYak_CAF1 100154 100 - 1 ----GUGUUGAAUGAAAUUGCGAG-GGCUUCUACUCAAAGUAAUUUACACUAAAUUACAUGCGAACAAGCAAAAUGAGAAAAGCCUACAAAGCUCUCGCUUAUGU ----...............(((((-(((((...((((..((((((((...)))))))).(((......)))...))))....(....).))))))))))...... ( -25.60) >DroAna_CAF1 95771 101 - 1 ----GCCUAAACUGAAAUUGCGAGGGGCUUCUACUCAAAGUAAUUUACACUAAAUUACAUGCGAACAAGCAAAAUGAGAAAAGCCUACAAAGCUCUCGCUUAUGG ----...............(((((((((((...((((..((((((((...)))))))).(((......)))...))))..))))))........)))))...... ( -24.90) >consensus ____GUGUUGAAUGAAAUUGCGAG_GGCUUCUACUCAAAGUAAUUUACACUAAAUUACAUGCGAACAAGCAAAAUGAGAAAAGCCUACAAAGCUCUCGCUUAUGU ...................(((((.(((((...((((..((((((((...)))))))).(((......)))...))))..))))).........)))))...... (-21.83 = -22.00 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:36 2006