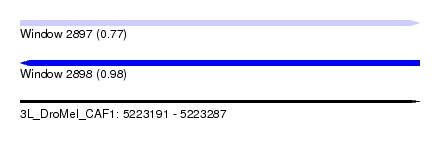
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,223,191 – 5,223,287 |
| Length | 96 |
| Max. P | 0.976742 |

| Location | 5,223,191 – 5,223,287 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.73 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -23.01 |
| Energy contribution | -23.43 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771966 |
| Prediction | RNA |
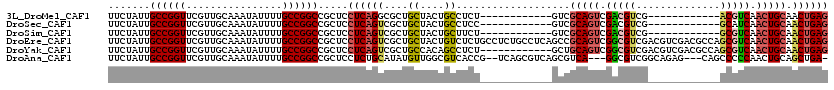
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5223191 96 + 23771897 UUCUAUUGCCGGUUCGUUGCAAAUAUUUUGCCGGCCGCUCCUCAGGCGCUGCUACUGCCUCU------------GUCGCAGUCGACGUCG------------ACGUCAACUGCAACUGAG .......((((((................))))))((((.....))))..((....))(((.------------((.(((((.((((...------------.)))).))))).)).))) ( -30.79) >DroSec_CAF1 87710 96 + 1 UUCUAUUGCCGGUUCGUUGCAAAUAUUUUGCCGGCCGCUCCUCAGUCGCUGCUACUGCCUCC------------GUCGCAGUCGACGUCG------------GCAUCAACUGCAACUGAG .......((((((................)))))).....((((((.((......((((..(------------((((....)))))..)------------)))......)).)))))) ( -33.19) >DroSim_CAF1 84811 96 + 1 UUCUAUUGCCGGUUCGUUGCAAAUAUUUUGCCGGCCGCUCCUCAGUCGCUGCUACUGCUUCU------------GUCGCAGUCGACGUCG------------GCGUCAACUGCAACUGAG .......((((((................)))))).....((((((....((....))....------------...(((((.((((...------------.)))).))))).)))))) ( -31.29) >DroEre_CAF1 89176 120 + 1 UUCUAUUGCCGGUUCGUUGCAAAUAUUUUGCCGGCCGCUCCUCAGUCGCUGCUACUGUCUCUGCCUCUGCCUCAGCCGCAGUCGGCGUCGACGUCGACGCCAGCGUCAACUGCAACUGAG .......((((((................)))))).....((((((.((((((.......((((..(((...)))..))))..(((((((....)))))))))))......)).)))))) ( -40.89) >DroYak_CAF1 89848 108 + 1 UUCUAUUGCCGGUUCGUUGCAAAUAUUUUGCCGGCCGCUCCUCAGUCGCUGCCACAGCCUCU------------GCUGCAGUCGGCGUCGACGUCGACGCCAGCGUCAACUGCAACUGAG .((..((((.((((((((((.........((((((.((....(((..((((...))))..))------------)..)).))))))((((....))))).)))))..))))))))..)). ( -39.70) >DroAna_CAF1 86751 111 + 1 UUCUAUUGCCGGUUCGUUGCAAAUAUUUUGCCGGCCGCUCCUCUGCAUAUGUUGGCGUCACCG--UCAGCGUCAGCGUCA---GGCGUCGGCAGAG---CAGCCCCCAACUGCAGCUGA- .((..((((.((((.(((((......(((((((((.(((....(((..(((((((((....))--)))))))..)))...---)))))))))))))---))))....))))))))..))- ( -39.20) >consensus UUCUAUUGCCGGUUCGUUGCAAAUAUUUUGCCGGCCGCUCCUCAGUCGCUGCUACUGCCUCU____________GCCGCAGUCGACGUCG____________GCGUCAACUGCAACUGAG .......((((((................)))))).....((((((....((....))...................(((((.(((((..............))))).))))).)))))) (-23.01 = -23.43 + 0.42)
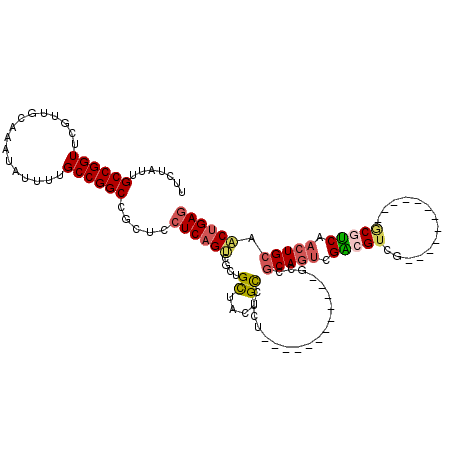
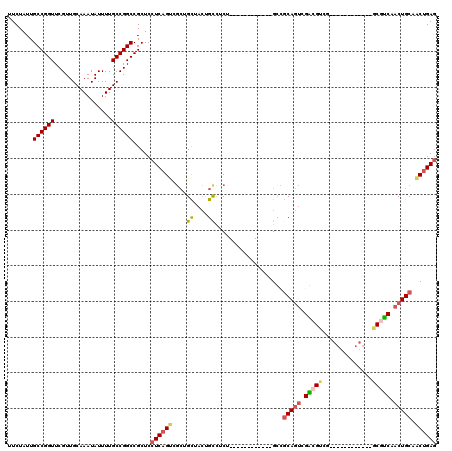
| Location | 5,223,191 – 5,223,287 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.73 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -23.86 |
| Energy contribution | -25.03 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5223191 96 - 23771897 CUCAGUUGCAGUUGACGU------------CGACGUCGACUGCGAC------------AGAGGCAGUAGCAGCGCCUGAGGAGCGGCCGGCAAAAUAUUUGCAACGAACCGGCAAUAGAA (((.(((((((((((((.------------...)))))))))))))------------..((((.((....))))))))).....(((((..................)))))....... ( -38.97) >DroSec_CAF1 87710 96 - 1 CUCAGUUGCAGUUGAUGC------------CGACGUCGACUGCGAC------------GGAGGCAGUAGCAGCGACUGAGGAGCGGCCGGCAAAAUAUUUGCAACGAACCGGCAAUAGAA (((((((((.((((.(((------------(..(((((....))))------------)..)))).)))).))))))))).....(((((..................)))))....... ( -43.27) >DroSim_CAF1 84811 96 - 1 CUCAGUUGCAGUUGACGC------------CGACGUCGACUGCGAC------------AGAAGCAGUAGCAGCGACUGAGGAGCGGCCGGCAAAAUAUUUGCAACGAACCGGCAAUAGAA (((((((((((((((((.------------...))))))))((.((------------.......)).)).))))))))).....(((((..................)))))....... ( -35.47) >DroEre_CAF1 89176 120 - 1 CUCAGUUGCAGUUGACGCUGGCGUCGACGUCGACGCCGACUGCGGCUGAGGCAGAGGCAGAGACAGUAGCAGCGACUGAGGAGCGGCCGGCAAAAUAUUUGCAACGAACCGGCAAUAGAA (((((((((.(((((((....)))))))(((...(((..((((.......)))).)))...))).......))))))))).....(((((..................)))))....... ( -48.77) >DroYak_CAF1 89848 108 - 1 CUCAGUUGCAGUUGACGCUGGCGUCGACGUCGACGCCGACUGCAGC------------AGAGGCUGUGGCAGCGACUGAGGAGCGGCCGGCAAAAUAUUUGCAACGAACCGGCAAUAGAA (((((((((..........(((((((....)))))))(.(..((((------------....))))..)).))))))))).....(((((..................)))))....... ( -47.37) >DroAna_CAF1 86751 111 - 1 -UCAGCUGCAGUUGGGGGCUG---CUCUGCCGACGCC---UGACGCUGACGCUGA--CGGUGACGCCAACAUAUGCAGAGGAGCGGCCGGCAAAAUAUUUGCAACGAACCGGCAAUAGAA -....(((..(((((.(((((---((((((....))(---((.(((....))((.--((....)).))......)))).))))))))).((((.....))))......)))))..))).. ( -34.90) >consensus CUCAGUUGCAGUUGACGC____________CGACGCCGACUGCGAC____________AGAGGCAGUAGCAGCGACUGAGGAGCGGCCGGCAAAAUAUUUGCAACGAACCGGCAAUAGAA (((.(((((((((((((................))))))))))))).................((((.......))))..)))..(((((..................)))))....... (-23.86 = -25.03 + 1.17)

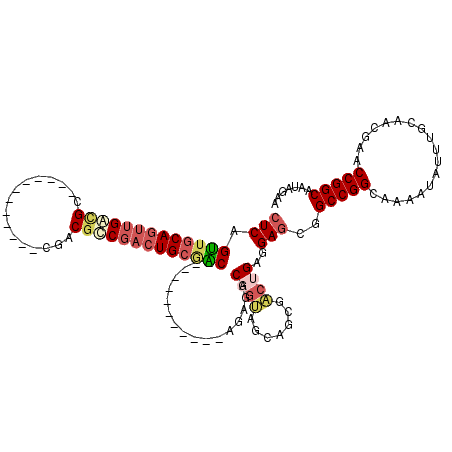
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:28 2006