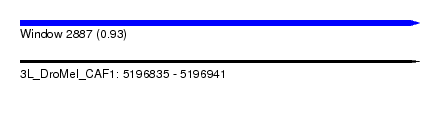
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,196,835 – 5,196,941 |
| Length | 106 |
| Max. P | 0.925200 |

| Location | 5,196,835 – 5,196,941 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.30 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -3.91 |
| Energy contribution | -4.11 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.17 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5196835 106 + 23771897 GCAAACAAAACUGGGACUCGGCACACGACU---CACUUGC-CAUGCAAAGGCACC-AAAAAACGACGAAGGUGGAAAAGCAUAAAACAAAAAAUAUUUGCAUUUUCCUCAA ...........(((((.(((.....))).)---)...(((-(.......))))))-).......((....))(((((((((................))).)))))).... ( -20.39) >DroVir_CAF1 74432 86 + 1 G---------------CGCUGCAGGCGGUU---CACUUGCCAAUGCAAAGUCC---AAAACAAGA--GCAGUGCAAAAGUAGAAUGCA--UAAUAUUUGCAUUCUGCUUAA (---------------((((((.(((((..---...)))))........((..---...))....--)))))))..((((((((((((--.......)))))))))))).. ( -34.10) >DroGri_CAF1 74556 92 + 1 G---------------UGCUGCAGGCGACUCGGCACUUGGCAAUGCAAAGACACAAAAACCAAGA--GCACUGCUAAAGUAGAAUGCA--UAAUAUUUGCAUACAGCUUAA (---------------(((((.((....)))))))).(((((.(((...................--))).)))))((((.(.(((((--.......))))).).)))).. ( -26.31) >DroSec_CAF1 61829 97 + 1 ---------ACUGGGACUCGGCACACGAUU---CACUUGC-CAUGCAAAGGCACC-AAAAAACGACGAAGGUGGAAAAACAUAAAACAAAAAAUAUUUGCAUUUUCCUCAA ---------..(((((.(((.....))).)---)...(((-(.......))))))-)......((.(((((((.(((..................))).))))))).)).. ( -18.77) >DroYak_CAF1 61903 82 + 1 GCAAACAAA---------------------------CUGC-AAUGCAAAGGCAUU-AAAAAAAGACGAAGGUGGAAAAGCAUAAAACAAAAAAUAUUUGCAUUUUCCUCAA (((......---------------------------.)))-(((((....)))))-........((....))(((((((((................))).)))))).... ( -14.39) >DroMoj_CAF1 82059 86 + 1 G---------------CGCUGCAAGUGAUU---CACUUGCCAAUGCAAAGACC---AAAACAAGA--GCAGUGCUAAAGUAGAAUGCA--UAAUAUUUGCAUUUUGCCUAA (---------------(((((((((((...---)))))((....)).......---.........--)))))))....((((((((((--.......)))))))))).... ( -27.00) >consensus G_______________CGCUGCACGCGAUU___CACUUGC_AAUGCAAAGGCAC__AAAAAAAGA__AAAGUGCAAAAGCAGAAAACA__AAAUAUUUGCAUUUUCCUCAA ....................................((((....))))........................(((((((((................))).)))))).... ( -3.91 = -4.11 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:18 2006