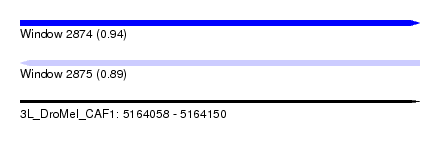
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,164,058 – 5,164,150 |
| Length | 92 |
| Max. P | 0.936099 |

| Location | 5,164,058 – 5,164,150 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -19.02 |
| Energy contribution | -18.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
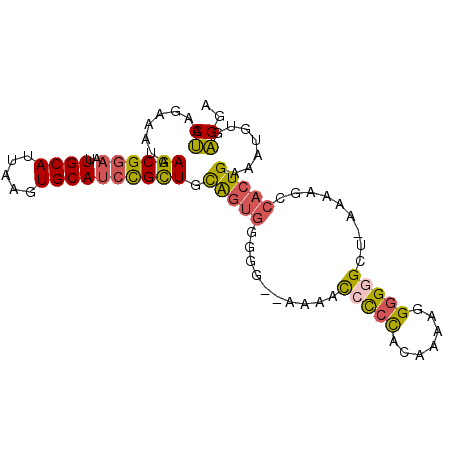
>3L_DroMel_CAF1 5164058 92 + 23771897 CUAAGAAAUAAGCGGAAUUGCAUUAAGUGCAUCCGCUGCAGUGGGGU--AAACCCCCCACAAAAAGGGGGCU-AAAAGCCACUGAAAUGUGAGGA ..........((((((..(((((...))))))))))).((((((...--....(((((.......)))))..-.....))))))........... ( -32.14) >DroSec_CAF1 29579 91 + 1 CUAAGAAAUAAGCGGAAUUGCAAUAAGUGCAUCCGCUGCAGUGUGGG--AAAAC-CCCACAAAAGGGGGGCU-AAAAGCCACUGAAAUGUGAGGA ..........((((((..((((.....)))))))))).(((((((((--.....-)))))........(((.-....)))))))........... ( -29.20) >DroSim_CAF1 29535 91 + 1 CUAAGAAAUAAGCGGAAUUGCAUUAAGUGCAUCCGCUGCAGUGUGGG--AAAAC-CCCACAAAAGGGGGGCU-AAAAGCCACUGAAAUGUGAGGA ..........((((((..(((((...))))))))))).(((((((((--.....-)))))........(((.-....)))))))........... ( -29.70) >DroEre_CAF1 27787 94 + 1 CUAAGAAAUAAGCGGAAUUGCAUUAAGUGCAUCUGCUGUAGUGGGGGGAAAAACCCCCACAAAAUGGGGGUU-AAAAGCCACUGAAAUGUGAGGA ..........((((((..(((((...)))))))))))...(((((((......)))))))....(.(((((.-....))).)).).......... ( -30.70) >DroYak_CAF1 27838 94 + 1 CUAAGAAAUAAGCGAAAUUGCAUUAAGUGCAGCUGUUGUAGUGGGGG-CAUAACCUCCACAAAAGGGGGGUUUCGAAGCCACUGAAUUGUGAGAA ....(((((..((((..((((((...))))))...)))).(((((((-.....))))))).........))))).....(((......))).... ( -24.20) >DroAna_CAF1 25740 80 + 1 CCAAGAAAUAAGCGGAAUUGCAUUAAGUGCAUCCGUUGCGA-G-GGG--C-AAUCUGUCCAAAAAGGGGG----------AAUGGAACGAGGGGG (((.......((((((..(((((...)))))))))))....-.-(((--(-.....))))..........----------..))).......... ( -18.40) >consensus CUAAGAAAUAAGCGGAAUUGCAUUAAGUGCAUCCGCUGCAGUGGGGG__AAAACCCCCACAAAAGGGGGGCU_AAAAGCCACUGAAAUGUGAGGA ((........((((((..((((.....)))))))))).(((((..........(((((.......))))).........))))).......)).. (-19.02 = -18.78 + -0.25)


| Location | 5,164,058 – 5,164,150 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -12.43 |
| Energy contribution | -15.68 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5164058 92 - 23771897 UCCUCACAUUUCAGUGGCUUUU-AGCCCCCUUUUUGUGGGGGGUUU--ACCCCACUGCAGCGGAUGCACUUAAUGCAAUUCCGCUUAUUUCUUAG ...........((((((.....-((((((((......)))))))).--...)))))).((((((((((.....))))..)))))).......... ( -35.00) >DroSec_CAF1 29579 91 - 1 UCCUCACAUUUCAGUGGCUUUU-AGCCCCCCUUUUGUGGG-GUUUU--CCCACACUGCAGCGGAUGCACUUAUUGCAAUUCCGCUUAUUUCUUAG ...........(((((......-((((((.(....).)))-)))..--....))))).((((((((((.....))))..)))))).......... ( -28.20) >DroSim_CAF1 29535 91 - 1 UCCUCACAUUUCAGUGGCUUUU-AGCCCCCCUUUUGUGGG-GUUUU--CCCACACUGCAGCGGAUGCACUUAAUGCAAUUCCGCUUAUUUCUUAG ...........(((((......-((((((.(....).)))-)))..--....))))).((((((((((.....))))..)))))).......... ( -28.10) >DroEre_CAF1 27787 94 - 1 UCCUCACAUUUCAGUGGCUUUU-AACCCCCAUUUUGUGGGGGUUUUUCCCCCCACUACAGCAGAUGCACUUAAUGCAAUUCCGCUUAUUUCUUAG ............(((((.....-(((((((((...))))))))).......)))))..(((.((((((.....))))..)).))).......... ( -25.80) >DroYak_CAF1 27838 94 - 1 UUCUCACAAUUCAGUGGCUUCGAAACCCCCCUUUUGUGGAGGUUAUG-CCCCCACUACAACAGCUGCACUUAAUGCAAUUUCGCUUAUUUCUUAG ............(((((...((.((((.(((....).)).)))).))-...))))).....(((((((.....)))).....))).......... ( -16.20) >DroAna_CAF1 25740 80 - 1 CCCCCUCGUUCCAUU----------CCCCCUUUUUGGACAGAUU-G--CCC-C-UCGCAACGGAUGCACUUAAUGCAAUUCCGCUUAUUUCUUGG .....((..((((..----------.........))))..))((-(--(..-.-..))))((((((((.....))))..))))............ ( -12.50) >consensus UCCUCACAUUUCAGUGGCUUUU_AGCCCCCCUUUUGUGGGGGUUUU__CCCCCACUGCAGCGGAUGCACUUAAUGCAAUUCCGCUUAUUUCUUAG ...........(((((.......(((((((.......)))))))........))))).((((((((((.....))))..)))))).......... (-12.43 = -15.68 + 3.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:06 2006