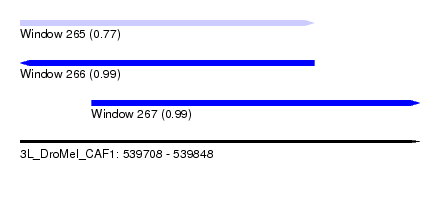
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 539,708 – 539,848 |
| Length | 140 |
| Max. P | 0.994419 |

| Location | 539,708 – 539,811 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.72 |
| Mean single sequence MFE | -39.95 |
| Consensus MFE | -23.81 |
| Energy contribution | -27.00 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 539708 103 + 23771897 GAAAUGUGGAC---------CCCCGCCCUCUCCUCCACCAACCUAUCUGCAAUUUGUGCUGGCGGAAAAGCGGGGAAAAGCGGGAAAAGCCGACGGA-AAGGGCAAACAUUCC ..(((((((..---------..))(((((.((((((.((.........(((.....)))..((......)))))))...((((......))).))))-.)))))..))))).. ( -30.80) >DroSec_CAF1 13738 112 + 1 GAAAUGUGCCCCCUCCUUCCCCCUGCCCUCCGCUCCACCAGCCCAUCUGCAAUUUGUGCAGGCGGGAAAAUGGGGAAAAGCGGGGAAAGCCGACGGA-AAGGGCAAACAUUCC (((...(((((..(((.((...((..((.(((((((.(((.(((..(((((.....)))))..)))....))))))...))))))..))..)).)))-..)))))....))). ( -46.00) >DroSim_CAF1 14647 112 + 1 GAAAUGUGCCCCCUCCUUCCCCCUGCCCUCCGCUCCACCAGCCCAUCUGCAAUUUGUGCAGGCGGGAAAAUGGGGAAAAGCGGGGAAAGCCGACGGA-AAGGGCAAACAUUCC (((...(((((..(((.((...((..((.(((((((.(((.(((..(((((.....)))))..)))....))))))...))))))..))..)).)))-..)))))....))). ( -46.00) >DroEre_CAF1 14151 98 + 1 GAAAUGUGGCCCUCCCUC--------------CUCCACCAGCCCUUCUGCAAUUUGUGCAGGCGGAAAAGCGGGG-AAAGCGGGGAAAGCCGGGGGAAAAGGGCAAACAUUCC ..(((((.(((((...((--------------((((.((..((((.(((((.....)))))((......))))))-.....))((....))))))))..)))))..))))).. ( -37.00) >consensus GAAAUGUGCCCCCUCCUU__CCCUGCCCUCCGCUCCACCAGCCCAUCUGCAAUUUGUGCAGGCGGAAAAACGGGGAAAAGCGGGGAAAGCCGACGGA_AAGGGCAAACAUUCC ..(((((.............((((((......((((((...(((..(((((.....)))))..)))...))))))....))))))...(((..........)))..))))).. (-23.81 = -27.00 + 3.19)

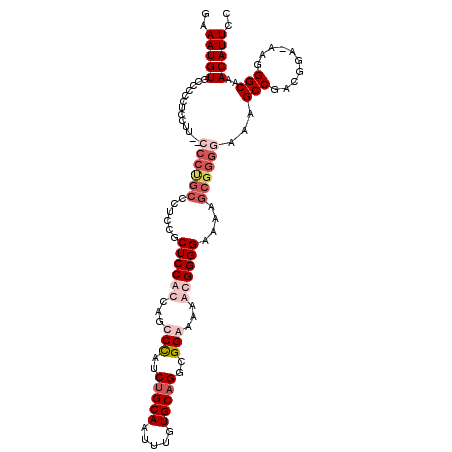
| Location | 539,708 – 539,811 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Mean single sequence MFE | -49.67 |
| Consensus MFE | -29.81 |
| Energy contribution | -35.12 |
| Covariance contribution | 5.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 539708 103 - 23771897 GGAAUGUUUGCCCUU-UCCGUCGGCUUUUCCCGCUUUUCCCCGCUUUUCCGCCAGCACAAAUUGCAGAUAGGUUGGUGGAGGAGAGGGCGGGG---------GUCCACAUUUC (((((((..((((..-.(((.(((......)))......(((.(((((((((((((...............))))))))))))).))))))))---------))..))))))) ( -40.36) >DroSec_CAF1 13738 112 - 1 GGAAUGUUUGCCCUU-UCCGUCGGCUUUCCCCGCUUUUCCCCAUUUUCCCGCCUGCACAAAUUGCAGAUGGGCUGGUGGAGCGGAGGGCAGGGGGAAGGAGGGGGCACAUUUC (((((((..((((((-((((....)((((((((((((((((((((..((((.(((((.....))))).))))..))))).).))))))).))))))))))))))))))))))) ( -58.50) >DroSim_CAF1 14647 112 - 1 GGAAUGUUUGCCCUU-UCCGUCGGCUUUCCCCGCUUUUCCCCAUUUUCCCGCCUGCACAAAUUGCAGAUGGGCUGGUGGAGCGGAGGGCAGGGGGAAGGAGGGGGCACAUUUC (((((((..((((((-((((....)((((((((((((((((((((..((((.(((((.....))))).))))..))))).).))))))).))))))))))))))))))))))) ( -58.50) >DroEre_CAF1 14151 98 - 1 GGAAUGUUUGCCCUUUUCCCCCGGCUUUCCCCGCUUU-CCCCGCUUUUCCGCCUGCACAAAUUGCAGAAGGGCUGGUGGAG--------------GAGGGAGGGCCACAUUUC (((((((..((((...((((.(((......)))...(-(((((((..(((..(((((.....)))))..)))..))))).)--------------)))))))))).))))))) ( -41.30) >consensus GGAAUGUUUGCCCUU_UCCGUCGGCUUUCCCCGCUUUUCCCCACUUUCCCGCCUGCACAAAUUGCAGAUGGGCUGGUGGAGCGGAGGGCAGGG__AAGGAGGGGCCACAUUUC (((((((..((((((..((...))(((((((((((((((((((((..((((.(((((.....))))).))))..))))).).))))))).))))))))..))))))))))))) (-29.81 = -35.12 + 5.31)

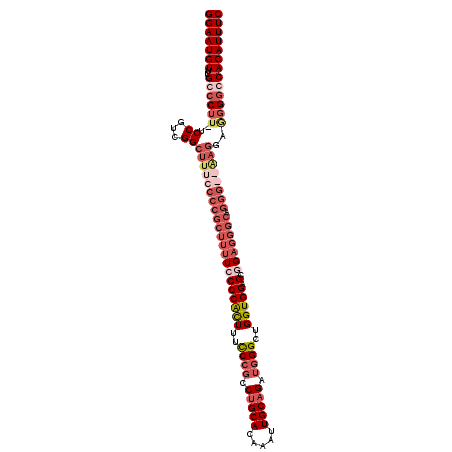
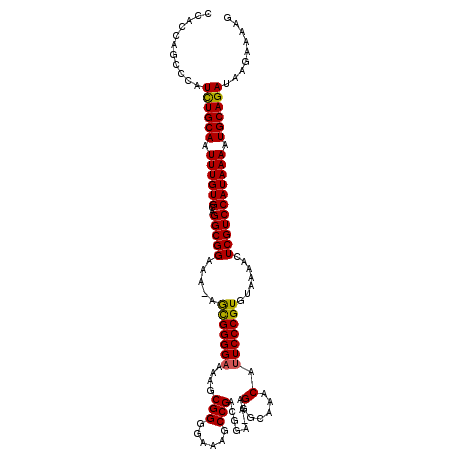
| Location | 539,733 – 539,848 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -28.80 |
| Energy contribution | -28.36 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 539733 115 + 23771897 CCACCAACCUAUCUGCAAUUUGUGCUGGCGGAAA-AGCGGGGAAAAGCGGGAAAAGCCGACGGA-AAGGGCAAACAUUCCCGUGUAAAACUCGUCCAUAAAAUGCAGAUAAGAAAAG .......(.((((((((.((((((...((.....-.))(((.....(((((((..(((......-...))).....)))))))......)))...)))))).)))))))).)..... ( -33.90) >DroSec_CAF1 13772 115 + 1 CCACCAGCCCAUCUGCAAUUUGUGCAGGCGGGAA-AAUGGGGAAAAGCGGGGAAAGCCGACGGA-AAGGGCAAACAUUCCCGUGUAAAACUCGUCCAUAAAAUGCAGAUAAGAAAAG ..........(((((((.((((((..((((((..-.(((((((...((((......))).)...-..(......).)))))))......)))))))))))).)))))))........ ( -32.30) >DroSim_CAF1 14681 115 + 1 CCACCAGCCCAUCUGCAAUUUGUGCAGGCGGGAA-AAUGGGGAAAAGCGGGGAAAGCCGACGGA-AAGGGCAAACAUUCCCGUGUAAAACUCGUCCAUAAAAUGCAGAUAAGAAAAG ..........(((((((.((((((..((((((..-.(((((((...((((......))).)...-..(......).)))))))......)))))))))))).)))))))........ ( -32.30) >DroEre_CAF1 14171 115 + 1 CCACCAGCCCUUCUGCAAUUUGUGCAGGCGGAAA-AGCGGGG-AAAGCGGGGAAAGCCGGGGGAAAAGGGCAAACAUUCCCGUGUAAAACUCGUCCAUAAAAUGCAGAUAAGAAAAG ...........((((((.((((((...((.....-.))(((.-...(((((((..(((..........))).....)))))))......)))...)))))).))))))......... ( -30.30) >DroYak_CAF1 14395 109 + 1 --------CCCUUUGCAAUUUGUGCAGGCGGAAUGGGCGGGGAAAAGCGGGGAAAGCCGGGGGAAAAGGGCAAACAUUCCCGUGUAAAACUCGUCCAUAAAAUGCAGAUAAGAAAAG --------((.((((((.....)))))).)).(((((((((.....(((((((..(((..........))).....)))))))......)))))))))................... ( -33.80) >consensus CCACCAGCCCAUCUGCAAUUUGUGCAGGCGGAAA_AGCGGGGAAAAGCGGGGAAAGCCGACGGA_AAGGGCAAACAUUCCCGUGUAAAACUCGUCCAUAAAAUGCAGAUAAGAAAAG ...........((((((.((((((..(((((.....(((((((....(((......)))........(......).))))))).......))))))))))).))))))......... (-28.80 = -28.36 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:13 2006