
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,161,166 – 5,161,290 |
| Length | 124 |
| Max. P | 0.874134 |

| Location | 5,161,166 – 5,161,265 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.47 |
| Mean single sequence MFE | -19.92 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.83 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5161166 99 - 23771897 UGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAUUAGUACUCUAAACAAAGAGUAUUACUGAAAA---UGU------------------ .((.((((...)))).))(((((.((((.((((((.....)))))).)))).)).)))....((.(((((((((......))))))))).))....---...------------------ ( -23.00) >DroSec_CAF1 24507 99 - 1 UGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGAUCAUCACAAUAGUACCUUAAACAAAGAAUAUUACCGAAAA---UGU------------------ (((((((...((.(((((((....(((((((.((....)).)))))))..))))))).))...)))))))..........................---...------------------ ( -16.10) >DroSim_CAF1 26655 99 - 1 UGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAAUAGUACUCUAAACAAAGAAUAUUGCCGAAAA---UGU------------------ (((((((...((.(((((((....(((((((.((....)).)))))))..))))))).))...))))))).............(((((......))---)))------------------ ( -15.70) >DroYak_CAF1 24935 120 - 1 UGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAAUAGUGCUGCAAACAAAGAACAGUAAUGAAAAACAUGUCAAAAACACACAUGAUAU ...(((((((((..((....(((.((((.((((((.....)))))).)))).)))(.((.(((....))).))).......))))))))))).....(((((.........))))).... ( -24.90) >consensus UGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAAUAGUACUCUAAACAAAGAAUAUUACCGAAAA___UGU__________________ (((((((...((.(((((((....(((((((.((....)).)))))))..))))))).))...))))))).................................................. (-14.95 = -14.83 + -0.12)


| Location | 5,161,185 – 5,161,290 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -20.80 |
| Energy contribution | -22.22 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5161185 105 + 23771897 UUGUUUAGAGUACUAAUGUGAUGACCGCAAUUGUUAAUAUUAAAUACAUUAUCAACGGCGUCGCGAUUACAGUAAUAGCAGCGACGCCGGCAGAAGCAUCCUCCC--- .(((((...((.....((((.....))))..........................(((((((((..(((......)))..)))))))))))..))))).......--- ( -25.00) >DroSec_CAF1 24526 105 + 1 UUGUUUAAGGUACUAUUGUGAUGAUCGCAAUUGUUAAUAUUAAAUACAUUAUCAACGGCGUCGCGAUUACAGUAAUAGCAGCGACGCCGGCAGAGGCAUCCUCCC--- .(((((((...((.((((((.....)))))).)).....))))))).........(((((((((..(((......)))..)))))))))...((((...))))..--- ( -31.90) >DroSim_CAF1 26674 105 + 1 UUGUUUAGAGUACUAUUGUGAUGACCGCAAUUGUUAAUAUUAAAUACAUUAUCAACGGCGUCGCGAUUACAGUAAUAGCAGCGACGCCGGCAGAGGCAUCCUCCC--- .(((((((...((.((((((.....)))))).)).....))))))).........(((((((((..(((......)))..)))))))))...((((...))))..--- ( -29.90) >DroEre_CAF1 24958 99 + 1 ------GCAGUACUAUUGAGAUGACCACAAUUGUUAAUAUUAAAUACAUUAUCAACGGCGUCGCGAUUACAGUAAUAGCAGCGACGCCGGCAGAGGCAUCCUCCC--- ------...........((((((.((....(((.((((.........)))).)))(((((((((..(((......)))..))))))))).....))))).)))..--- ( -26.30) >DroYak_CAF1 24975 105 + 1 UUGUUUGCAGCACUAUUGUGAUGACCGCAAUUGUUAAUAUUAAAUACAUUAUCAACGGCGUCGCGAUUACAGUAAUAGCAGCGACGCCGGCAGAAGCAUCCUCCC--- ...(((((((((..((((((.....))))))))))....................(((((((((..(((......)))..))))))))))))))...........--- ( -30.30) >DroAna_CAF1 23546 84 + 1 ----------------UGUGAUA--------CGUGUAUAUAAAAUACAUUAUCAGCGGCGUCGCGAUUACAGUAAUAGCAGCGACGCCGGCAGCGGCAUCCUCUUUUC ----------------..(((((--------.((((((.....))))))))))).(((((((((..(((......)))..)))))))))((....))........... ( -27.30) >consensus UUGUUUACAGUACUAUUGUGAUGACCGCAAUUGUUAAUAUUAAAUACAUUAUCAACGGCGUCGCGAUUACAGUAAUAGCAGCGACGCCGGCAGAGGCAUCCUCCC___ ...........((.((((((.....)))))).)).....................(((((((((..(((......)))..)))))))))((....))........... (-20.80 = -22.22 + 1.42)


| Location | 5,161,185 – 5,161,290 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -25.32 |
| Energy contribution | -26.27 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
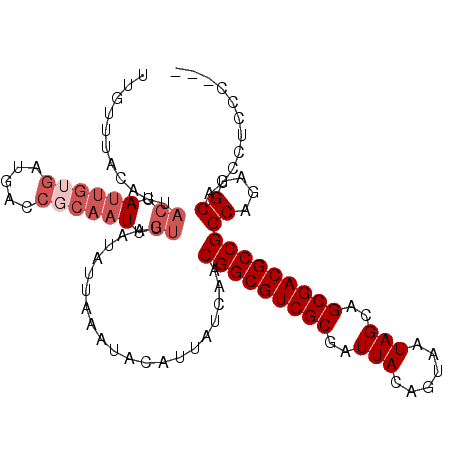
>3L_DroMel_CAF1 5161185 105 - 23771897 ---GGGAGGAUGCUUCUGCCGGCGUCGCUGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAUUAGUACUCUAAACAA ---.((((((((...((((.(((((((((((.......)))...))))))))(((((((.((....)).)))))))....)))).))))((....)).))))...... ( -33.40) >DroSec_CAF1 24526 105 - 1 ---GGGAGGAUGCCUCUGCCGGCGUCGCUGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGAUCAUCACAAUAGUACCUUAAACAA ---..((((.(((.......(((((((((((.......)))...))))))))(((((((.((....)).))))))).((((.(.....).)))).)))))))...... ( -28.70) >DroSim_CAF1 26674 105 - 1 ---GGGAGGAUGCCUCUGCCGGCGUCGCUGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAAUAGUACUCUAAACAA ---.((((((((...((((.(((((((((((.......)))...))))))))(((((((.((....)).)))))))....)))).))))((....)).))))...... ( -33.40) >DroEre_CAF1 24958 99 - 1 ---GGGAGGAUGCCUCUGCCGGCGUCGCUGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGUGGUCAUCUCAAUAGUACUGC------ ---..(((.(((((.(....(((((((((((.......)))...))))))))(((((((.((....)).)))))))....).)).))))))...........------ ( -29.80) >DroYak_CAF1 24975 105 - 1 ---GGGAGGAUGCUUCUGCCGGCGUCGCUGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAAUAGUGCUGCAAACAA ---((.((((...)))).))(((((((((((.......)))...))))))))(((((((.((....)).)))))))..((((((.(((.......))))))))).... ( -34.60) >DroAna_CAF1 23546 84 - 1 GAAAAGAGGAUGCCGCUGCCGGCGUCGCUGCUAUUACUGUAAUCGCGACGCCGCUGAUAAUGUAUUUUAUAUACACG--------UAUCACA---------------- ........(((((....((.(((((((((((.......)))...))))))))))......(((((.....))))).)--------))))...---------------- ( -26.00) >consensus ___GGGAGGAUGCCUCUGCCGGCGUCGCUGCUAUUACUGUAAUCGCGACGCCGUUGAUAAUGUAUUUAAUAUUAACAAUUGCGGUCAUCACAAUAGUACUCUAAACAA ........((((...((((((((((((((((.......)))...)))))))))(((.((((((.....)))))).)))..)))).))))................... (-25.32 = -26.27 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:03 2006