
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,160,715 – 5,160,808 |
| Length | 93 |
| Max. P | 0.896356 |

| Location | 5,160,715 – 5,160,808 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -18.92 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.48 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
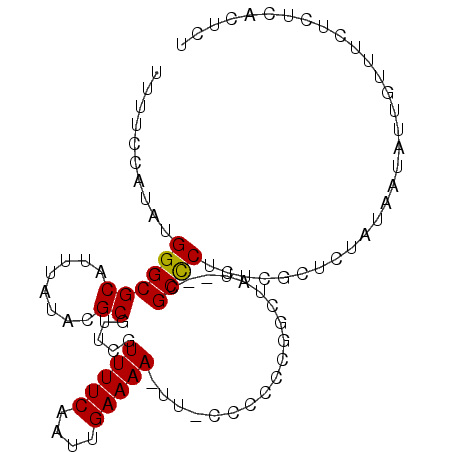
>3L_DroMel_CAF1 5160715 93 + 23771897 AGAG--AGAGAAACAAUAUUAUAGAGCGAGAGGGCG--AUAGCCGUGGGG-AA-UUUUCAAUUGAAAACGAACGCGUAUAAAUGCGCCCAUAUGGAAAA ....--....................(....)(((.--...)))(((((.-..-(((((....)))))....(((((....))))))))))........ ( -18.60) >DroVir_CAF1 31370 84 + 1 -GACUGACAGCAG--AC--GAG----GGAGAGGGCGCGGCA----AGCGG-AA-UUUUCAAUUGAAAACGAAUGCGUAUAAAUGCGCCCAUAUGGAAAA -..(((....)))--..--...----.....(((((((...----.(((.-..-(((((....)))))....))).......))))))).......... ( -21.10) >DroEre_CAF1 24492 94 + 1 AGAGUGAGAGAAACAAUAUUAUAGAGCGAGAGGGCG--AUAGCCGG-GGG-AA-UUUUCAAUUGAAAACGAACGCGUAUAAAUGCGCCCAUAUGGAAAA ..................(((((.........(((.--...)))((-(..-..-(((((....)))))....(((((....)))))))).))))).... ( -17.00) >DroWil_CAF1 33438 92 + 1 ----UGAGUGUGUGGGUGUGCGACAGCGAGCGAGCG--AGAGAAAGGCGG-AAUUUUUCAAUUGAAAAUGAAUGCGUAUAAAUGCGCCCAUAUGGAAAA ----.....((((((((((......((......)).--........(((.-...(((((....)))))....)))........))))))))))...... ( -23.10) >DroYak_CAF1 24476 94 + 1 AGAGUGAGAGAAACAAUAUUAAAGAGCGAGAGGGCG--AUAGGCGG-GGG-AA-UUUUCAAUUGAAAACGAACGCGUAUAAAUGCGCCCAUAUGGAAAA ...............................(((((--....(((.-(..-..-(((((....)))))....).))).......))))).......... ( -17.70) >DroAna_CAF1 23198 96 + 1 AGAGAGAGAGAGAGAAUAUAGAAGAGGGAGAGGGCU--AUAGCCGGGGGAAAA-UUUUCAAUUGAAAACGAACGCGUAUAAAUGCGCCCAUAUGGAAAA ................................(((.--...)))(((......-(((((....)))))....(((((....)))))))).......... ( -16.00) >consensus AGAGUGAGAGAAACAAUAUUAAAGAGCGAGAGGGCG__AUAGCCGGGGGG_AA_UUUUCAAUUGAAAACGAACGCGUAUAAAUGCGCCCAUAUGGAAAA ...............................((((...................(((((....))))).....((((....)))))))).......... (-12.32 = -12.48 + 0.17)



| Location | 5,160,715 – 5,160,808 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -14.12 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.62 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5160715 93 - 23771897 UUUUCCAUAUGGGCGCAUUUAUACGCGUUCGUUUUCAAUUGAAAA-UU-CCCCACGGCUAU--CGCCCUCUCGCUCUAUAAUAUUGUUUCUCU--CUCU ..........((((((........))))))((((((....)))))-).-......(((...--.)))..........................--.... ( -14.10) >DroVir_CAF1 31370 84 - 1 UUUUCCAUAUGGGCGCAUUUAUACGCAUUCGUUUUCAAUUGAAAA-UU-CCGCU----UGCCGCGCCCUCUCC----CUC--GU--CUGCUGUCAGUC- ..........((((((........(((..(((((((....)))))-..-.))..----))).)))))).....----...--(.--(((....))).)- ( -16.40) >DroEre_CAF1 24492 94 - 1 UUUUCCAUAUGGGCGCAUUUAUACGCGUUCGUUUUCAAUUGAAAA-UU-CCC-CCGGCUAU--CGCCCUCUCGCUCUAUAAUAUUGUUUCUCUCACUCU ..........((((((........))))))((((((....)))))-).-...-..(((...--.)))................................ ( -14.10) >DroWil_CAF1 33438 92 - 1 UUUUCCAUAUGGGCGCAUUUAUACGCAUUCAUUUUCAAUUGAAAAAUU-CCGCCUUUCUCU--CGCUCGCUCGCUGUCGCACACCCACACACUCA---- ..........((((((........)).....(((((....)))))...-..))))......--.((..((.....)).))...............---- ( -10.80) >DroYak_CAF1 24476 94 - 1 UUUUCCAUAUGGGCGCAUUUAUACGCGUUCGUUUUCAAUUGAAAA-UU-CCC-CCGCCUAU--CGCCCUCUCGCUCUUUAAUAUUGUUUCUCUCACUCU ..........(((((.....(((.(((...((((((....)))))-).-...-.))).)))--)))))............................... ( -14.90) >DroAna_CAF1 23198 96 - 1 UUUUCCAUAUGGGCGCAUUUAUACGCGUUCGUUUUCAAUUGAAAA-UUUUCCCCCGGCUAU--AGCCCUCUCCCUCUUCUAUAUUCUCUCUCUCUCUCU ..........((((((........))))))((((((....)))))-)........(((...--.)))................................ ( -14.40) >consensus UUUUCCAUAUGGGCGCAUUUAUACGCGUUCGUUUUCAAUUGAAAA_UU_CCCCCCGGCUAU__CGCCCUCUCGCUCUAUAAUAUUGUUUCUCUCACUCU ..........((((((........)).....(((((....)))))...................))))............................... (-10.76 = -10.62 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:01 2006