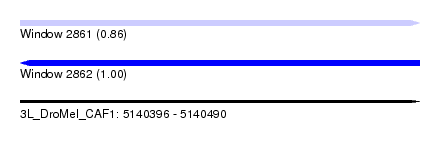
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,140,396 – 5,140,490 |
| Length | 94 |
| Max. P | 0.999627 |

| Location | 5,140,396 – 5,140,490 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -11.28 |
| Energy contribution | -12.52 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5140396 94 + 23771897 UUUUGACGCCUUCUGAGUUUGUAAGUUUGAGAAUAUGAAUAUUCUCAGGAAGUGGAAACCCACUUAAU-UUUUAGGCUCCUGUAUCUAUGCAUAU .......((((...(((((......((((((((((....))))))))))((((((....)))))))))-))..))))...((((....))))... ( -23.60) >DroSec_CAF1 3945 72 + 1 UUUUGGCGCCUCCUUAGGUUGUAAGUUUGAGAAUAUGAAUAUUCUCAGAAAGUGGAAGCCCACUUAAU-UUGU---------------------- .....((((((....))).)))...((((((((((....))))))))))((((((....))))))...-....---------------------- ( -19.40) >DroSim_CAF1 2826 72 + 1 UUUUGGCGCCUCCUUAGGUUGUAAGUUUGAGAAUAUGAAUAUUCUCAGGAAGUGGAAGCCCACUUAAU-UUGU---------------------- .....((((((....))).)))...((((((((((....))))))))))((((((....))))))...-....---------------------- ( -18.70) >DroEre_CAF1 3894 92 + 1 UUUUGGCGCCUCCUUAGGAUAUAAUUUUGAGAAUAUGAC--UUCUCAGGAAGUCGGAACCCAAUUAAU-UUGUGGGCUGCUGUAUCUAUGCAUUC ....(((((((.((((((((...))))))))....((((--(((....))))))).............-....)))).))).............. ( -19.80) >DroYak_CAF1 3914 90 + 1 UUUUGGCACCUCAU---AAUAUAAGUUUGAGAAUAUAAC--UUCUCAGGAAGUGGGAACCCAAUUAAUUUUGUAGGCUCCUGUUUCUAUGCAUAU .....(((......---..........((((((......--))))))(((((..(((.(((((......)))..)).)))..))))).))).... ( -21.60) >consensus UUUUGGCGCCUCCUUAGGUUGUAAGUUUGAGAAUAUGAAUAUUCUCAGGAAGUGGAAACCCACUUAAU_UUGU_GGCU_CUGU_UCUAUGCAU__ .........................((((((((((....))))))))))((((((....)))))).............................. (-11.28 = -12.52 + 1.24)

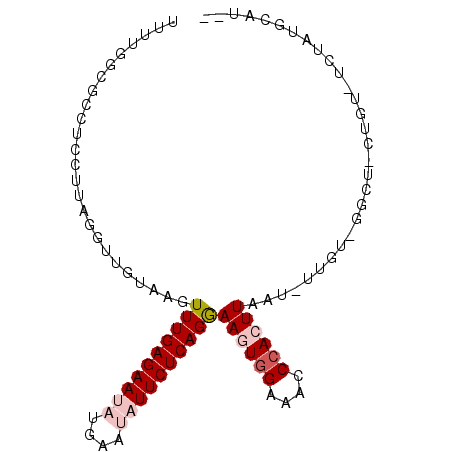
| Location | 5,140,396 – 5,140,490 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -19.74 |
| Consensus MFE | -8.80 |
| Energy contribution | -10.20 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5140396 94 - 23771897 AUAUGCAUAGAUACAGGAGCCUAAAA-AUUAAGUGGGUUUCCACUUCCUGAGAAUAUUCAUAUUCUCAAACUUACAAACUCAGAAGGCGUCAAAA ...............(..((((....-...((((((....))))))..((((((((....))))))))................))))..).... ( -21.30) >DroSec_CAF1 3945 72 - 1 ----------------------ACAA-AUUAAGUGGGCUUCCACUUUCUGAGAAUAUUCAUAUUCUCAAACUUACAACCUAAGGAGGCGCCAAAA ----------------------....-......(((((((((......((((((((....))))))))...(((.....))))))))).)))... ( -19.10) >DroSim_CAF1 2826 72 - 1 ----------------------ACAA-AUUAAGUGGGCUUCCACUUCCUGAGAAUAUUCAUAUUCUCAAACUUACAACCUAAGGAGGCGCCAAAA ----------------------....-......(((((((((......((((((((....))))))))...(((.....))))))))).)))... ( -19.10) >DroEre_CAF1 3894 92 - 1 GAAUGCAUAGAUACAGCAGCCCACAA-AUUAAUUGGGUUCCGACUUCCUGAGAA--GUCAUAUUCUCAAAAUUAUAUCCUAAGGAGGCGCCAAAA ....((.........(.((((((...-......)))))).)(.(((((((((((--......))))))...(((.....)))))))))))..... ( -18.30) >DroYak_CAF1 3914 90 - 1 AUAUGCAUAGAAACAGGAGCCUACAAAAUUAAUUGGGUUCCCACUUCCUGAGAA--GUUAUAUUCUCAAACUUAUAUU---AUGAGGUGCCAAAA ....((((.(((...((((((((..........))))))))...))).((((((--......))))))..(((((...---)))))))))..... ( -20.90) >consensus __AUGCAUAGA_ACAG_AGCC_ACAA_AUUAAGUGGGUUUCCACUUCCUGAGAAUAUUCAUAUUCUCAAACUUACAACCUAAGGAGGCGCCAAAA ..............................((((((....))))))..((((((((....))))))))........................... ( -8.80 = -10.20 + 1.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:54 2006