
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,126,150 – 5,126,292 |
| Length | 142 |
| Max. P | 0.847755 |

| Location | 5,126,150 – 5,126,265 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.84 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5126150 115 + 23771897 CCGGCCAGGUCGAUGGCCACUUUCUUUCCCCGCAGUUCAUUAAUGGGUUUACGUUCACAAUGGGGCGUUAAAACACCCCAUAAUUCCUUGACGCCCAUUUA-AAUAAGCUAAACAA ..(((((......)))))...............((((.((((((((((...........((((((.((....)).))))))...........)))))))..-))).))))...... ( -30.85) >DroVir_CAF1 3788 113 + 1 CCGGCCAGAUCUAUGGCGACCUUCUUGCCGCGGAGUUCACAUAUUGGCUUUCGCUCAGCAUGUGGCGUUAAGAUGCUCCACAGUUCCUUAACGCCCAUUUA-AU-UAU-UAAACAC ..((((((.((..((((((.....))))))..))(....)...))))))..........(((.(((((((((..(((....)))..))))))))))))...-..-...-....... ( -32.10) >DroEre_CAF1 3676 115 + 1 CCGGCCAGGUCAAUGGCCACUUUCUUUCCGCGCAGCUCAUUAAUAGGCUUACGUUCACAAUGGGGCGUCAAAACUCCCCAUAAUUCCUUGACGCCCAUUUA-AAUGAACUAAAUAA ..(((((......)))))...............((((........))))...(((((.....(((((((((................))))))))).....-..)))))....... ( -31.19) >DroYak_CAF1 3706 115 + 1 CCGGCCAGGUCAAUAGCAACUUUCUUUCCUCGCAGUUCGUUAAUGGGUUUACGUUCACAAUGGGGCGUCAAAACUCCCCACAAUUCCUUGACGCCCAUUUA-AAUAAACUAAAUAA ...((.(((..................))).))((((..(((((((((............(((((.((....)).)))))............)))))))..-))..))))...... ( -23.82) >DroAna_CAF1 3684 116 + 1 CCGGCCAGAUCGAUGGCCACCUUCUUGCCUCGCAGCUCACUAAUGGGCUUCCGCUCGCAGUGAGGGGUUAAAACAGUCCAUAGCUCCUUAACACCCAUUCAAAAUAAAAUAAAUAA ..(((((......))))).(((((((((..((.((((((....))))))..))...)))).)))))(((((...(((.....)))..)))))........................ ( -30.00) >DroPer_CAF1 2420 113 + 1 CCGGCCAAAUCAAUGGCCACUUUCUUGCCGCGAAGCUCAUUUAUCGGCUUGCGCUCAGCAUGAGGGGUUAAAACUGUCCAAAGUUCUUUUACACCCAUAAA--ACAAU-GAAACAA ..(((((......))))).(((...(((.((((((((........))))).)))...))).)))(((((((((((......)))...)))).)))).....--.....-....... ( -28.50) >consensus CCGGCCAGAUCAAUGGCCACUUUCUUGCCGCGCAGCUCAUUAAUGGGCUUACGCUCACAAUGGGGCGUUAAAACUCCCCAUAAUUCCUUGACGCCCAUUUA_AAUAAACUAAACAA ..(((((......)))))...............((((........))))..........(((.((((((((................))))))))))).................. (-15.78 = -15.84 + 0.06)

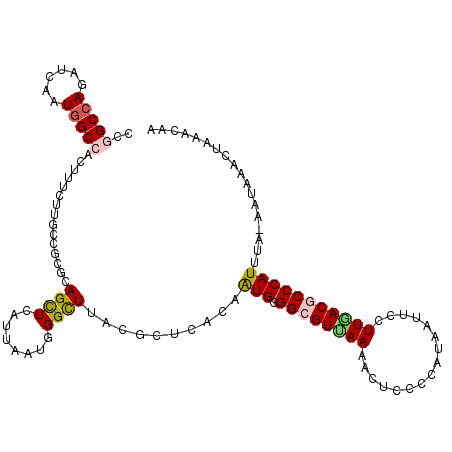
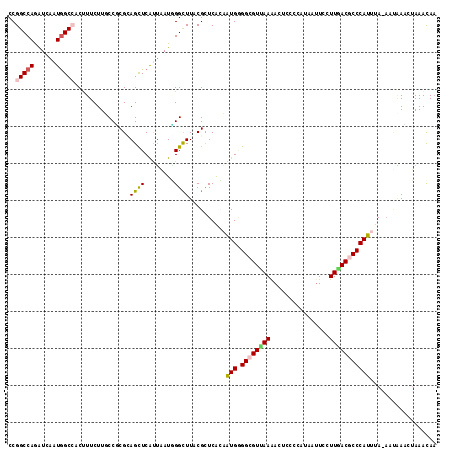
| Location | 5,126,190 – 5,126,292 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -14.86 |
| Energy contribution | -16.00 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5126190 102 + 23771897 UAAUGGGUUUACGUUCACAAUGGGGCGUUAAAACACCCCAUAAUUCCUUGACGCCCAUUUAAAUAAGCUAAACAAAAACC--CCGCGAGAAAUUCGCAACAAAU ....((((((.(((.....)))(((((((((................)))))))))...................)))))--).(((((...)))))....... ( -24.19) >DroVir_CAF1 3828 101 + 1 AUAUUGGCUUUCGCUCAGCAUGUGGCGUUAAGAUGCUCCACAGUUCCUUAACGCCCAUUUAAU-UAU-UAAACACUAAAUUAGCGCGUUAAUUUCAAAACGAG- ..((((((...(((.....(((.(((((((((..(((....)))..)))))))))))).((((-(..-.........)))))))).))))))...........- ( -21.50) >DroSec_CAF1 3721 102 + 1 UAAUGGGUUUGCGUUCACAAUGGGGCGUCAAAACACCCCAUAAUUCCUUGACGCCCAUUUAAAUAAGCUAGACAGAAACC--CCGCGAAAUAUUCGCAACAAAU ....((((((((..........(((((((((................)))))))))..........)).......)))))--).((((.....))))....... ( -27.05) >DroSim_CAF1 3750 102 + 1 UAAUGGGCUUGCGUUCACAAUGGGGCGUCAAAACACCCCAUAAUUCCUUUACGCCCAUUUAAAUAAGCUAAACAGAAACC--CCGCGAGAAAUCCGCAACAAAU .(((((((...........((((((.((....)).))))))...........))))))).....................--..((((....).)))....... ( -24.05) >DroEre_CAF1 3716 102 + 1 UAAUAGGCUUACGUUCACAAUGGGGCGUCAAAACUCCCCAUAAUUCCUUGACGCCCAUUUAAAUGAACUAAAUAAAAACC--CCGCGCGAAAUUCGCAACAAAU ............(((((.....(((((((((................))))))))).......)))))............--....(((.....)))....... ( -23.59) >DroYak_CAF1 3746 102 + 1 UAAUGGGUUUACGUUCACAAUGGGGCGUCAAAACUCCCCACAAUUCCUUGACGCCCAUUUAAAUAAACUAAAUAAAAACC--CCGCGAGAAAUUAUCAACAAAU ....((((((.(((.....)))(((((((((................)))))))))...................)))))--)(....)............... ( -22.29) >consensus UAAUGGGCUUACGUUCACAAUGGGGCGUCAAAACACCCCAUAAUUCCUUGACGCCCAUUUAAAUAAGCUAAACAAAAACC__CCGCGAGAAAUUCGCAACAAAU .(((((((...........((((((.((....)).))))))...........))))))).........................(((.......)))....... (-14.86 = -16.00 + 1.14)

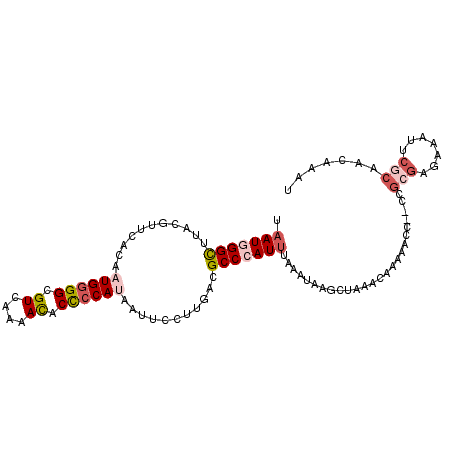
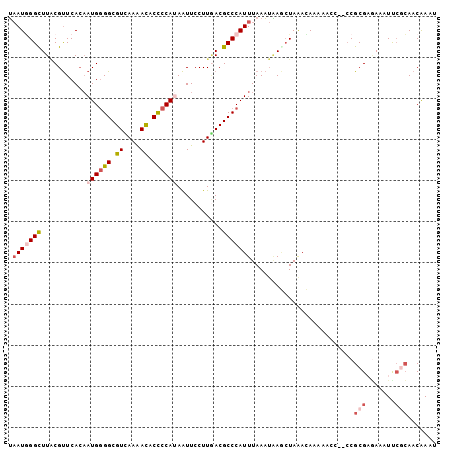
| Location | 5,126,190 – 5,126,292 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -18.46 |
| Energy contribution | -19.63 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5126190 102 - 23771897 AUUUGUUGCGAAUUUCUCGCGG--GGUUUUUGUUUAGCUUAUUUAAAUGGGCGUCAAGGAAUUAUGGGGUGUUUUAACGCCCCAUUGUGAACGUAAACCCAUUA ......(((((.....)))))(--(((((.((((((((((((....))))))...........(((((((((....)))))))))..)))))).)))))).... ( -36.70) >DroVir_CAF1 3828 101 - 1 -CUCGUUUUGAAAUUAACGCGCUAAUUUAGUGUUUA-AUA-AUUAAAUGGGCGUUAAGGAACUGUGGAGCAUCUUAACGCCACAUGCUGAGCGAAAGCCAAUAU -..(((((....(((((.((((((...)))))))))-)).-...)))))((((((((((..((....))..))))))))))......((.((....)))).... ( -27.60) >DroSec_CAF1 3721 102 - 1 AUUUGUUGCGAAUAUUUCGCGG--GGUUUCUGUCUAGCUUAUUUAAAUGGGCGUCAAGGAAUUAUGGGGUGUUUUGACGCCCCAUUGUGAACGCAAACCCAUUA .......((((.....))))((--(((((((.....((((((....))))))....))))))..((((((((....))))))))((((....)))).))).... ( -33.70) >DroSim_CAF1 3750 102 - 1 AUUUGUUGCGGAUUUCUCGCGG--GGUUUCUGUUUAGCUUAUUUAAAUGGGCGUAAAGGAAUUAUGGGGUGUUUUGACGCCCCAUUGUGAACGCAAGCCCAUUA ......(((((.....)))))(--(((((.((((((((((((....))))))...........(((((((((....)))))))))..)))))).)))))).... ( -36.00) >DroEre_CAF1 3716 102 - 1 AUUUGUUGCGAAUUUCGCGCGG--GGUUUUUAUUUAGUUCAUUUAAAUGGGCGUCAAGGAAUUAUGGGGAGUUUUGACGCCCCAUUGUGAACGUAAGCCUAUUA ...(((.(((.....))))))(--(((((.......((((((......(((((((((((..((....))..)))))))))))....))))))..)))))).... ( -31.10) >DroYak_CAF1 3746 102 - 1 AUUUGUUGAUAAUUUCUCGCGG--GGUUUUUAUUUAGUUUAUUUAAAUGGGCGUCAAGGAAUUGUGGGGAGUUUUGACGCCCCAUUGUGAACGUAAACCCAUUA .....................(--(((((.......((((((......(((((((((((..(......)..)))))))))))....))))))..)))))).... ( -26.60) >consensus AUUUGUUGCGAAUUUCUCGCGG__GGUUUUUGUUUAGCUUAUUUAAAUGGGCGUCAAGGAAUUAUGGGGUGUUUUGACGCCCCAUUGUGAACGUAAACCCAUUA ......(((((.....)))))...(((((.((((((.........((((((((((((((..((....))..)))))))))).)))).)))))).)))))..... (-18.46 = -19.63 + 1.17)

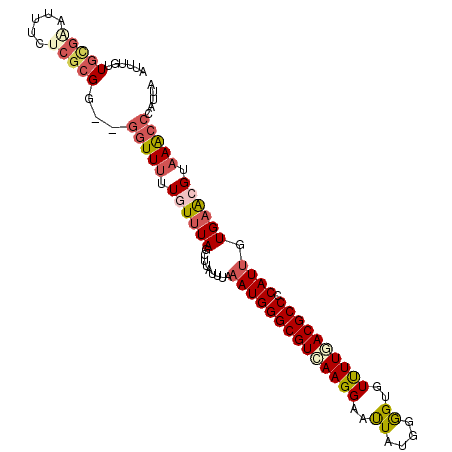
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:49 2006