
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,118,380 – 5,118,483 |
| Length | 103 |
| Max. P | 0.863760 |

| Location | 5,118,380 – 5,118,483 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
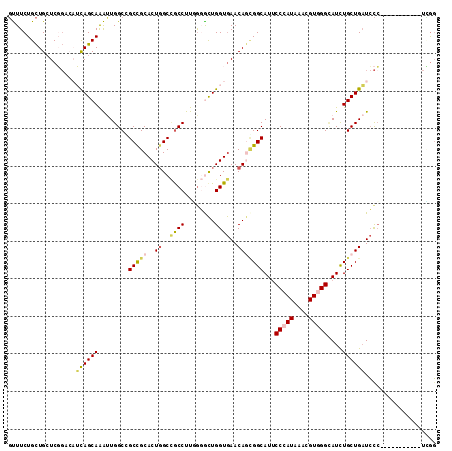
>3L_DroMel_CAF1 5118380 103 + 23771897 GUUUCUGCUGAUCGGACAUCAGCAAAUUGGCGGCUGCACUGGCCGCCUUGGGGCUGGUGAACAGCGGCAUUCCCAUAAACGUGGGUAUCUGCUGAUCCC-----------UUGG ((..((((((((.....)))))))....(((((((.....)))))))..)..)).((.((.((((((.((.(((((....))))).)))))))).))))-----------.... ( -45.90) >DroVir_CAF1 4764 114 + 1 GCUUCUGCUGCUCCGCCUGCAGCAGGCUGGCCGCCGCACUCGCUGCCUUCACGUUGGUAAACAAGGGCAUGCCCAUGAAAGUAGGUAUUUGCUGCUGCGCCGACGACGACUGAC ((..(((((((.......))))))))).(((.((.......)).)))....(((((((...((.(((....))).))..(((((.......)))))..)))))))......... ( -39.30) >DroSec_CAF1 4637 103 + 1 GUUUCUGCUGCUCGGUCAUCAGCAAAUUGGCCGCUGCACUGGCCGCCUUGGGGCUGGUGAACAGCGGCAUUCCCAUAAACGUGGGCAUCUGCUGAUCCC-----------UCGG ............(((..(((((((...((((((((((((..(((.(....))))..)))..)))))))...(((((....)))))))..)))))))..)-----------.)). ( -44.50) >DroSim_CAF1 4627 103 + 1 GUUUCUGCUGCUCGGUCAUCAGCAAAUUGGCCGCUGCACUGGCCGCCUUGGGGCUGGUGAACAGCGGCAUUCCCAUAAACGUGGGCAUCUGCUGAUCCC-----------UCGG ............(((..(((((((...((((((((((((..(((.(....))))..)))..)))))))...(((((....)))))))..)))))))..)-----------.)). ( -44.50) >DroEre_CAF1 4620 103 + 1 GUUUCUGCUGCUCGGAGAUCAGCAAACUGGCGGCCGCACUGGCCGCCUUUGGACUGGUGAACAGCGGCAUUCCCAUGAACGUAGGCAUCUGCUGAUCCC-----------UCGG .....(((((((.(...((((((.....(((((((.....)))))))....).)))))...))))))))..........((.(((.(((....))).))-----------))). ( -38.60) >DroAna_CAF1 5402 97 + 1 UUUUCUGUUGCUC------UAGCAGAUUGGCGGCCGCACUGGCAACCUUUGCACUGGUGAACAAUGGCAUUCCCAUAAAGGUGGGAAUCUGCUGAUCUC-----------UAGG ............(------(((.((((..(((((((((((.(((.....)))...)))).....))))((((((((....)))))))).)))..)))))-----------))). ( -34.20) >consensus GUUUCUGCUGCUCGGACAUCAGCAAAUUGGCCGCCGCACUGGCCGCCUUGGGGCUGGUGAACAGCGGCAUUCCCAUAAACGUGGGCAUCUGCUGAUCCC___________UCGG ..................((((((........(((((..((..((((........))))..)))))))...(((((....)))))....))))))................... (-24.10 = -24.68 + 0.59)


| Location | 5,118,380 – 5,118,483 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -25.41 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5118380 103 - 23771897 CCAA-----------GGGAUCAGCAGAUACCCACGUUUAUGGGAAUGCCGCUGUUCACCAGCCCCAAGGCGGCCAGUGCAGCCGCCAAUUUGCUGAUGUCCGAUCAGCAGAAAC ....-----------(((....(((....((((......))))..))).((((.....)))))))..((((((.......))))))..(((((((((.....)))))))))... ( -40.30) >DroVir_CAF1 4764 114 - 1 GUCAGUCGUCGUCGGCGCAGCAGCAAAUACCUACUUUCAUGGGCAUGCCCUUGUUUACCAACGUGAAGGCAGCGAGUGCGGCGGCCAGCCUGCUGCAGGCGGAGCAGCAGAAGC ((..((((((((...(((.((................((.(((....))).))(((((....))))).)).)))...))))))))..))(((((((.......))))))).... ( -39.20) >DroSec_CAF1 4637 103 - 1 CCGA-----------GGGAUCAGCAGAUGCCCACGUUUAUGGGAAUGCCGCUGUUCACCAGCCCCAAGGCGGCCAGUGCAGCGGCCAAUUUGCUGAUGACCGAGCAGCAGAAAC .((.-----------(..((((((((((.((((......))))...(((((((..(((..((((....).)))..))))))))))..))))))))))..)))............ ( -42.00) >DroSim_CAF1 4627 103 - 1 CCGA-----------GGGAUCAGCAGAUGCCCACGUUUAUGGGAAUGCCGCUGUUCACCAGCCCCAAGGCGGCCAGUGCAGCGGCCAAUUUGCUGAUGACCGAGCAGCAGAAAC .((.-----------(..((((((((((.((((......))))...(((((((..(((..((((....).)))..))))))))))..))))))))))..)))............ ( -42.00) >DroEre_CAF1 4620 103 - 1 CCGA-----------GGGAUCAGCAGAUGCCUACGUUCAUGGGAAUGCCGCUGUUCACCAGUCCAAAGGCGGCCAGUGCGGCCGCCAGUUUGCUGAUCUCCGAGCAGCAGAAAC .((.-----------(((((((((((((......(..(.(((((((......)))).))))..)...(((((((.....))))))).)))))))))))))))............ ( -44.50) >DroAna_CAF1 5402 97 - 1 CCUA-----------GAGAUCAGCAGAUUCCCACCUUUAUGGGAAUGCCAUUGUUCACCAGUGCAAAGGUUGCCAGUGCGGCCGCCAAUCUGCUA------GAGCAACAGAAAA .(((-----------(((((..((..(((((((......)))))))((.((((.....))))))...((((((....))))))))..)))).)))------)............ ( -29.50) >consensus CCGA___________GGGAUCAGCAGAUGCCCACGUUUAUGGGAAUGCCGCUGUUCACCAGCCCCAAGGCGGCCAGUGCAGCCGCCAAUUUGCUGAUGACCGAGCAGCAGAAAC ...............((..(((((((((.((((......))))......((((.....)))).....(((.((.......)).))).)))))))))...))............. (-25.41 = -24.80 + -0.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:45 2006