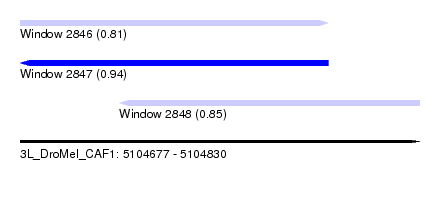
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,104,677 – 5,104,830 |
| Length | 153 |
| Max. P | 0.936452 |

| Location | 5,104,677 – 5,104,795 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -21.91 |
| Energy contribution | -22.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5104677 118 + 23771897 GUUU--UCAGCUGCAAGAUCGUCGUCAGCAUCAUCAAUACUUGCACGAUGACUGGGCAAAAAAAACACUUGGCCUCCGUACCAAUCAUAUGUUGGUUUUUAUACCAUCGCCCAGCCCAAA ....--............(((((((..(((...........))))))))))((((((...........((((........))))......(.((((......)))).)))))))...... ( -27.40) >DroSec_CAF1 297592 118 + 1 GUUUUUUUAGCUGCAAGAUCGUCGUUAUCAUCAUCGAUACUUGCACGAUGACUGGGCAAA--AAACACUUGGCCUCCGUACCAAUCAUAGGUUGGUUUUUAUACCAUCGCCCAGUCCAAA .........(.((((((((((..(.......)..)))).)))))))...((((((((...--......((((........)))).....(((.(((......)))))))))))))).... ( -34.00) >DroSim_CAF1 319270 118 + 1 GUUUUUUUACCUGCAAGAUAGUCGUCAUAAUCAUCGAUACUUGCACGAUGACUGGGCAAA--AAACACUUGGCCUCCGUACCAAUCAUAGGUUGGUUUUUAUACCAUCGCCCAGCCCAAA .........(.((((((...((((..........)))).)))))).).((.((((((...--......((((........)))).....(((.(((......))))))))))))..)).. ( -27.50) >DroYak_CAF1 309937 113 + 1 GUUU--UCAGCUGCAAGAUCAUCU---CCAUCGUCGAUACUUGCACGAUGACUGGGCUGA--AAACACUUGGCCUCCGUACCAUUCAUAGAUUGGUUUUUAUACCACAGCCCCACCCAAA (((.--((.(.(((((((((..(.---.....)..))).))))))))).))).((((((.--.......(((........))).........((((......))))))))))........ ( -27.50) >consensus GUUU__UCAGCUGCAAGAUCGUCGUCAUCAUCAUCGAUACUUGCACGAUGACUGGGCAAA__AAACACUUGGCCUCCGUACCAAUCAUAGGUUGGUUUUUAUACCAUCGCCCAGCCCAAA ......(((.(((((((...((((..........)))).)))))).).)))((((((............(((........))).........((((......))))..))))))...... (-21.91 = -22.47 + 0.56)



| Location | 5,104,677 – 5,104,795 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -33.42 |
| Energy contribution | -32.80 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5104677 118 - 23771897 UUUGGGCUGGGCGAUGGUAUAAAAACCAACAUAUGAUUGGUACGGAGGCCAAGUGUUUUUUUUGCCCAGUCAUCGUGCAAGUAUUGAUGAUGCUGACGACGAUCUUGCAGCUGA--AAAC ....((((((((((((((......))))((((....(((((......))))))))).....)))))))))).(((((((((.((((.((.......)).))))))))))..)))--.... ( -35.70) >DroSec_CAF1 297592 118 - 1 UUUGGACUGGGCGAUGGUAUAAAAACCAACCUAUGAUUGGUACGGAGGCCAAGUGUUU--UUUGCCCAGUCAUCGUGCAAGUAUCGAUGAUGAUAACGACGAUCUUGCAGCUAAAAAAAC ....((((((((((((((......))))........(((((......)))))......--.))))))))))....((((((.((((.((.......)).))))))))))........... ( -37.90) >DroSim_CAF1 319270 118 - 1 UUUGGGCUGGGCGAUGGUAUAAAAACCAACCUAUGAUUGGUACGGAGGCCAAGUGUUU--UUUGCCCAGUCAUCGUGCAAGUAUCGAUGAUUAUGACGACUAUCUUGCAGGUAAAAAAAC ....((((((((((((((......))))........(((((......)))))......--.))))))))))..(.(((((((((((..........))).)).)))))).)......... ( -34.90) >DroYak_CAF1 309937 113 - 1 UUUGGGUGGGGCUGUGGUAUAAAAACCAAUCUAUGAAUGGUACGGAGGCCAAGUGUUU--UCAGCCCAGUCAUCGUGCAAGUAUCGACGAUGG---AGAUGAUCUUGCAGCUGA--AAAC (((.(((.((((((((((......)))).........((((......)))).......--.))))))........((((((.((((.(.....---.).))))))))))))).)--)).. ( -32.20) >consensus UUUGGGCUGGGCGAUGGUAUAAAAACCAACCUAUGAUUGGUACGGAGGCCAAGUGUUU__UUUGCCCAGUCAUCGUGCAAGUAUCGAUGAUGAUGACGACGAUCUUGCAGCUAA__AAAC ....((((((((((((((......))))........(((((......))))).........))))))))))....((((((.((((.((.......)).))))))))))........... (-33.42 = -32.80 + -0.62)

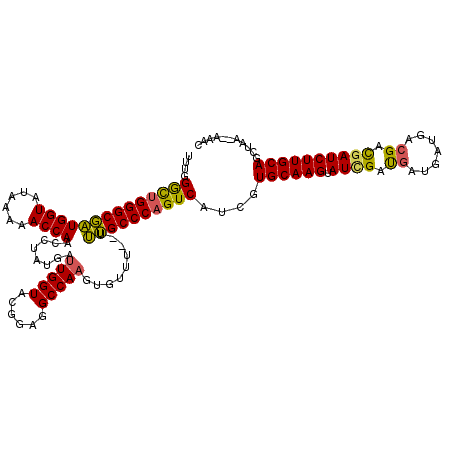

| Location | 5,104,715 – 5,104,830 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -27.34 |
| Energy contribution | -26.78 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5104715 115 - 23771897 AAAUCAAUAAAAGUUA-----AGCUAAACGACGGAACCAUUUUGGGCUGGGCGAUGGUAUAAAAACCAACAUAUGAUUGGUACGGAGGCCAAGUGUUUUUUUUGCCCAGUCAUCGUGCAA ................-----.((...((((((((.....))))((((((((((((((......))))((((....(((((......))))))))).....)))))))))).)))))).. ( -30.60) >DroSec_CAF1 297632 113 - 1 AAAUCAAUAAAAGUAA-----AGCUAAAGGACGGAACCAUUUUGGACUGGGCGAUGGUAUAAAAACCAACCUAUGAUUGGUACGGAGGCCAAGUGUUU--UUUGCCCAGUCAUCGUGCAA ................-----.......(.((((...(.....)((((((((((((((......))))........(((((......)))))......--.)))))))))).)))).).. ( -29.60) >DroSim_CAF1 319310 113 - 1 AAAUCAAUAAAAGUAA-----AGCUAAACGACGGAACCAUUUUGGGCUGGGCGAUGGUAUAAAAACCAACCUAUGAUUGGUACGGAGGCCAAGUGUUU--UUUGCCCAGUCAUCGUGCAA ................-----.((...((((((((.....))))((((((((((((((......))))........(((((......)))))......--.)))))))))).)))))).. ( -30.20) >DroYak_CAF1 309972 118 - 1 GAAUCAAUAAAAGUAAAGUUGAGCUAAGCGAUGGAACCAUUUUGGGUGGGGCUGUGGUAUAAAAACCAAUCUAUGAAUGGUACGGAGGCCAAGUGUUU--UCAGCCCAGUCAUCGUGCAA ...(((((.........)))))((...(((((((.(((......))).((((((((((......)))).........((((......)))).......--.))))))..))))))))).. ( -31.90) >consensus AAAUCAAUAAAAGUAA_____AGCUAAACGACGGAACCAUUUUGGGCUGGGCGAUGGUAUAAAAACCAACCUAUGAUUGGUACGGAGGCCAAGUGUUU__UUUGCCCAGUCAUCGUGCAA ......................((...((((.(....)......((((((((((((((......))))........(((((......))))).........)))))))))).)))))).. (-27.34 = -26.78 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:41 2006