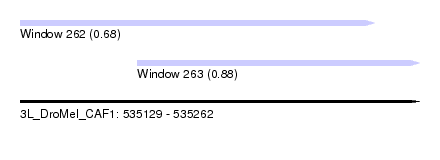
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 535,129 – 535,262 |
| Length | 133 |
| Max. P | 0.877001 |

| Location | 535,129 – 535,247 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -27.72 |
| Energy contribution | -27.24 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 535129 118 + 23771897 CACAAUCGAUGGCAGAAAUGAAAUUGU-UUUUUCACAACUCAAAACUCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAG .......(((..((....))..)))((-((((((((..(((.......)))....((...(((((....(((((((.((....))))))))).))))).)).....))))))).))).. ( -34.50) >DroSec_CAF1 9224 117 + 1 CACAAUCAAUGGCAGAAACGAAAUCGU-U-UUUCACAACUCAAAACUCGGGAAGGUCAAACUGGGGUUGGGUGGGUACUUAAAGGGUCCGCCACUCAGGGAAACUGGUGAAAAUGACAG .........((..(((((((....)))-)-)))..)).(((.......)))...((((..(((((....((((((..(.....)..)))))).)))))(....).........)))).. ( -29.70) >DroSim_CAF1 10089 117 + 1 CACAAUCAAUGGCAGAAACGAAAUCGU-U-UUUCACAACUCAAAACUCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAG .........(((((((((((....)))-)-))).....(((.......)))...))))..(((((....(((((((.((....))))))))).)))))(....)((((....))))... ( -34.40) >DroEre_CAF1 9514 119 + 1 CGCCAUCGAUGGCAGAAAUACAAUUUUUUUUUUCACAACUCAAAACUCGGGAAGGUCAAAUUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAG .((((....))))................(((((((..(((.......)))....((...(((((....(((((((.((....))))))))).))))).)).....)))))))...... ( -33.30) >DroYak_CAF1 9658 118 + 1 CUCAAUCAAUGGGAGAAAUAAAAUUGU-GUUUUCACAACUCAAAACGCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAG .....(((.(((((....)....((((-(....)))))))))...((((((....((...(((((....(((((((.((....))))))))).))))).))..))))))....)))... ( -36.20) >consensus CACAAUCAAUGGCAGAAAUGAAAUUGU_UUUUUCACAACUCAAAACUCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAG ..............................((((((..(((.......)))..(((....(((((....(((((((.((....))))))))).)))))....))).))))))....... (-27.72 = -27.24 + -0.48)

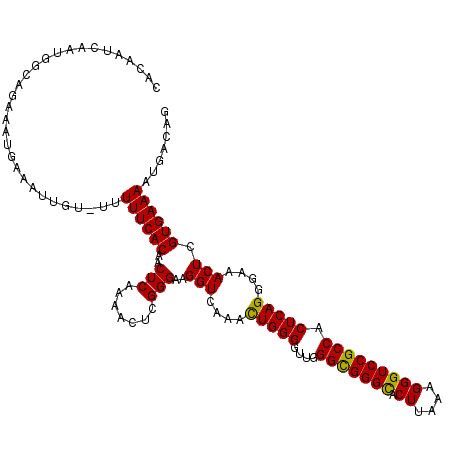
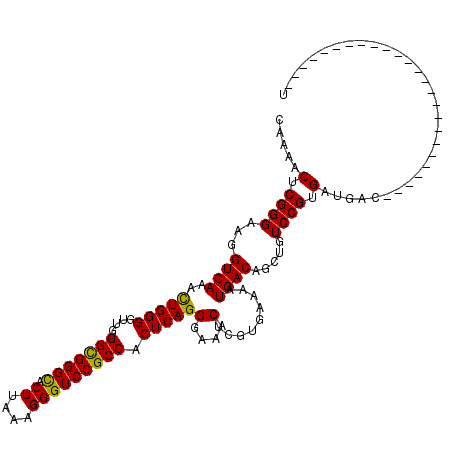
| Location | 535,168 – 535,262 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.32 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 535168 94 + 23771897 CAAAACUCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAGCUGUCCGUGAUGAC-------------------------U .....(.((((...((((..(((((....(((((((.((....))))))))).)))))(....).........)))).....)))).).....-------------------------. ( -32.50) >DroSec_CAF1 9262 94 + 1 CAAAACUCGGGAAGGUCAAACUGGGGUUGGGUGGGUACUUAAAGGGUCCGCCACUCAGGGAAACUGGUGAAAAUGACAGCUGUCCGUGAUGAC-------------------------U .....(.((((...((((..(((((....((((((..(.....)..)))))).)))))(....).........)))).....)))).).....-------------------------. ( -28.10) >DroSim_CAF1 10127 94 + 1 CAAAACUCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAGCUGUCCGUGAUGAC-------------------------U .....(.((((...((((..(((((....(((((((.((....))))))))).)))))(....).........)))).....)))).).....-------------------------. ( -32.50) >DroEre_CAF1 9554 101 + 1 CAAAACUCGGGAAGGUCAAAUUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAGCUGUCCGUGAUGACUGUA------------------GACU ......((.....(((((...(((((((((((((((.((....)))))))))......(....)((((....)))))))))..)))...)))))...------------------)).. ( -30.60) >DroYak_CAF1 9697 119 + 1 CAAAACGCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAGCUGUCCGUGAUGACUGCGGACUGUAGACUGUGUGUUGACU (((.((((((....((((..(((((....(((((((.((....))))))))).)))))(....).........)))).((.((((((.......)))))).))...)))))).)))... ( -46.30) >consensus CAAAACUCGGGAAGGUCAAACUGGGGUUGGGCGGGCACUUAAAGGGUCCGCCACUCAGGGAAACUCGUGAAAAUGACAGCUGUCCGUGAUGAC_________________________U .....(.((((...((((..(((((....(((((((.((....))))))))).)))))(....).........)))).....)))).)............................... (-30.80 = -30.32 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:08 2006