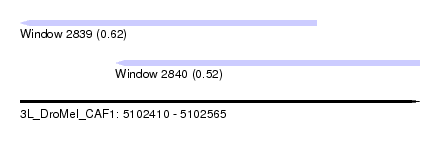
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,102,410 – 5,102,565 |
| Length | 155 |
| Max. P | 0.617433 |

| Location | 5,102,410 – 5,102,525 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -25.35 |
| Energy contribution | -26.35 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5102410 115 - 23771897 AACAGAGAUAGCAAACAUGGCGCAAAAAGUGUACUUUGUUUGCAUAUGUCUGACCUUUUGCA-UCGCACUCGGCACGUG--UUGCGACUAUCGAAAGAGGGCAGGUGACUA--AAACGAA ..((((.((((((((((..((((.....))))....))))))).))).)))).((..(((((-..((((.......)))--))))))...((....))))..((....)).--....... ( -34.40) >DroSec_CAF1 295432 116 - 1 AACAGAGCUAGCAAACAUGGGGCAAAAGGCGUACUUUGUUUGCAUAUGUCUGUCCUUUUGAACUCGCACUCGGCACGUG--UUGCGACUAUCGAAAAAAGGCAGGUGACUA--AAACGAA ..........(((((((.((.((.....))...)).)))))))...(..(((((.((((((..((((((.((...)).)--.)))))...))))))...)))))..)....--....... ( -28.00) >DroSim_CAF1 317042 115 - 1 AACAGAGAUAGCAAACAUGGCGCAAAAAGCGUACUUUGUUUGCAUAUGUCUGACCUUUUGCACUCGCACUCGGCACGUG--UUGCGACUAUCGAAA-AAGGCAGGUGACUA--AAAUGAA ..((((.((((((((((..((((.....))))....))))))).))).)))).((((((....((((((.((...)).)--.))))).......))-)))).((....)).--....... ( -35.20) >DroYak_CAF1 307716 119 - 1 AAGAGAGAUAGCAAACAUGGCGCAAAAAGCGUACUUUGUUUGCAUAUGUGUGACCUUUCGCA-UCGCACUUGGCACGUGUGUGGCGACUAUCGAAAAAAGGCAGUUGCCUAUGAAACGAA ..........(((((((..((((.....))))....)))))))(((..((((.((....((.-..))....))))))..)))(((((((..(.......)..)))))))........... ( -36.60) >consensus AACAGAGAUAGCAAACAUGGCGCAAAAAGCGUACUUUGUUUGCAUAUGUCUGACCUUUUGCA_UCGCACUCGGCACGUG__UUGCGACUAUCGAAAAAAGGCAGGUGACUA__AAACGAA ..((((.((((((((((..((((.....))))....))))))).))).)))).((((((....((((.....)).(((.....)))......)).)))))).((....)).......... (-25.35 = -26.35 + 1.00)

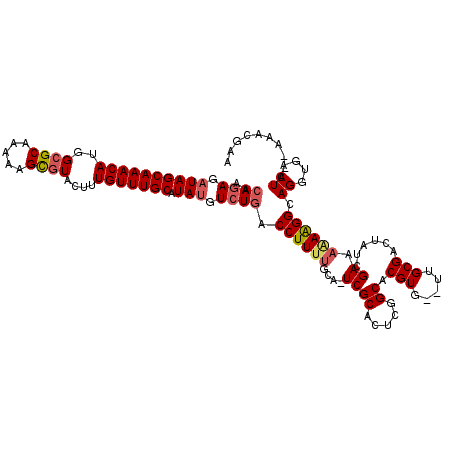
| Location | 5,102,447 – 5,102,565 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.73 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -27.25 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5102447 118 - 23771897 UGGGCGGCUUAAAGGGGGUGUACAUAGUGGAUAAAGGGGGAACAGAGAUAGCAAACAUGGCGCAAAAAGUGUACUUUGUUUGCAUAUGUCUGACCUUUUGCA-UCGCACUCGGCACGUG- ...((((((......((((((.(.....)(((.((((((...((((.((((((((((..((((.....))))....))))))).))).)))).))))))..)-))))))))))).))).- ( -37.80) >DroSec_CAF1 295469 119 - 1 UGGGCGGCUUAAAGGGGGUGUGCACAAUGGAUAAAGGGGGAACAGAGCUAGCAAACAUGGGGCAAAAGGCGUACUUUGUUUGCAUAUGUCUGUCCUUUUGAACUCGCACUCGGCACGUG- ...((((((......(((((((......(..(((((((...(((((..(((((((((.((.((.....))...)).))))))).))..))))))))))))..).)))))))))).))).- ( -34.00) >DroSim_CAF1 317078 119 - 1 UGGGCGGCUUAAAGGGGGUGUGCACAAUGGAUAAGGGGGGAACAGAGAUAGCAAACAUGGCGCAAAAAGCGUACUUUGUUUGCAUAUGUCUGACCUUUUGCACUCGCACUCGGCACGUG- ...((((((......(((((((......(..(((((((....((((.((((((((((..((((.....))))....))))))).))).)))).)))))))..).)))))))))).))).- ( -40.60) >DroYak_CAF1 307756 109 - 1 UGGGUG----------GUUGUACACCAUGGAUGGAGGGGGAAGAGAGAUAGCAAACAUGGCGCAAAAAGCGUACUUUGUUUGCAUAUGUGUGACCUUUCGCA-UCGCACUUGGCACGUGU ..((((----------......))))...((((((((((..(.(.(.((.(((((((..((((.....))))....))))))))).).).)..)))))).))-))((((.......)))) ( -36.70) >consensus UGGGCGGCUUAAAGGGGGUGUACACAAUGGAUAAAGGGGGAACAGAGAUAGCAAACAUGGCGCAAAAAGCGUACUUUGUUUGCAUAUGUCUGACCUUUUGCA_UCGCACUCGGCACGUG_ ...................((((....((((..((((((...((((.((((((((((..((((.....))))....))))))).))).)))).))))))....)).))....)))).... (-27.25 = -28.00 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:33 2006