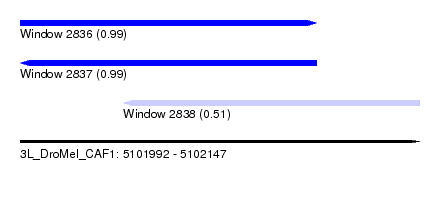
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,101,992 – 5,102,147 |
| Length | 155 |
| Max. P | 0.993595 |

| Location | 5,101,992 – 5,102,107 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -30.05 |
| Energy contribution | -29.68 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5101992 115 + 23771897 CAACCGGUUCUGCAUUAAAAUGAUCUUGCUGAUCUUCUGGUGCAGCAGCAGCAGCAACUUCUUCUUCAUUUUGCCUCAAUUUGGCAAUAUGGACGAAGCAACCACA-----AUAUAGCCG ....(((((((((((((.((.((((.....)))))).))))))))..((....)).....(((((((((.(((((.......))))).))))).))))........-----....))))) ( -32.50) >DroSec_CAF1 295033 109 + 1 CAACCGGUUCUGCAGCAAAAUGAUCUUGCUGAUCUUCUGGUGCAGC------AGCAACUUCUUCUUCGUUUUGCCUCAAUUUGGAAAUAUGGAAGAAGCAACCACA-----UUAUAGCCG ....((((((((((.((.((.((((.....)))))).)).))))).------.....((((((((..(((...((.......)).)))..))))))))........-----....))))) ( -28.70) >DroSim_CAF1 316647 112 + 1 CAACCGGUUCUGCAGCAAAAUGAUCUUGCUGAUCUUCUGGUGCAGCA---GCAGCAACUUCUUCUUCAUUUUGCCUCAAUUUGGCAAUAUGGAAGAAGCAACCACA-----AUAUAGCCG ....((((((((((.((.((.((((.....)))))).)).)))))..---.......(((((((..(((.(((((.......))))).))))))))))........-----....))))) ( -33.80) >DroYak_CAF1 307317 114 + 1 CAACCGGUUCUGCAGCAAAAUGAUCUUGCUGAUCUUCUGGUGCAGC------AGCAGCUUCUUCUUCGUUUUGCCUCAAUUUGGCAAUAUGAAAGAAGAAGCCACAUAGCCAUAUAGCCG ....((((((((((.((.((.((((.....)))))).)).))))).------....(((((((((((((.(((((.......))))).))).)))))))))).............))))) ( -41.30) >consensus CAACCGGUUCUGCAGCAAAAUGAUCUUGCUGAUCUUCUGGUGCAGC______AGCAACUUCUUCUUCAUUUUGCCUCAAUUUGGCAAUAUGGAAGAAGCAACCACA_____AUAUAGCCG ....((((((((((.((.((.((((.....)))))).)).)))))...............(((((((((.(((((.......))))).))))).)))).................))))) (-30.05 = -29.68 + -0.37)


| Location | 5,101,992 – 5,102,107 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -32.44 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5101992 115 - 23771897 CGGCUAUAU-----UGUGGUUGCUUCGUCCAUAUUGCCAAAUUGAGGCAAAAUGAAGAAGAAGUUGCUGCUGCUGCUGCACCAGAAGAUCAGCAAGAUCAUUUUAAUGCAGAACCGGUUG (((......-----.(..((.(((((.(((((.(((((.......))))).)))..)).))))).))..).....(((((..((((((((.....)))).))))..)))))..))).... ( -34.30) >DroSec_CAF1 295033 109 - 1 CGGCUAUAA-----UGUGGUUGCUUCUUCCAUAUUUCCAAAUUGAGGCAAAACGAAGAAGAAGUUGCU------GCUGCACCAGAAGAUCAGCAAGAUCAUUUUGCUGCAGAACCGGUUG (((......-----.(..((.((((((((....(((((.......)).))).....)))))))).)).------.)((((.(((((((((.....)))).))))).))))...))).... ( -30.60) >DroSim_CAF1 316647 112 - 1 CGGCUAUAU-----UGUGGUUGCUUCUUCCAUAUUGCCAAAUUGAGGCAAAAUGAAGAAGAAGUUGCUGC---UGCUGCACCAGAAGAUCAGCAAGAUCAUUUUGCUGCAGAACCGGUUG (((......-----.(..((.(((((((((((.(((((.......))))).)))..)))))))).))..)---..(((((.(((((((((.....)))).))))).)))))..))).... ( -41.90) >DroYak_CAF1 307317 114 - 1 CGGCUAUAUGGCUAUGUGGCUUCUUCUUUCAUAUUGCCAAAUUGAGGCAAAACGAAGAAGAAGCUGCU------GCUGCACCAGAAGAUCAGCAAGAUCAUUUUGCUGCAGAACCGGUUG (((............(..(((((((((((....(((((.......)))))...)))))))))))..).------.(((((.(((((((((.....)))).))))).)))))..))).... ( -43.50) >consensus CGGCUAUAU_____UGUGGUUGCUUCUUCCAUAUUGCCAAAUUGAGGCAAAACGAAGAAGAAGUUGCU______GCUGCACCAGAAGAUCAGCAAGAUCAUUUUGCUGCAGAACCGGUUG (((............(..(((.(((((((....(((((.......)))))...))))))).)))..)........(((((.(((((((((.....)))).))))).)))))..))).... (-32.44 = -32.62 + 0.19)



| Location | 5,102,032 – 5,102,147 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -19.11 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5102032 115 - 23771897 UUUGUUUUUAUCGUCUGAAAUUUACAAUUCAAUGUCAUGCCGGCUAUAU-----UGUGGUUGCUUCGUCCAUAUUGCCAAAUUGAGGCAAAAUGAAGAAGAAGUUGCUGCUGCUGCUGCA ...........(((.((((........)))))))...(((.(((.....-----.(..((.(((((.(((((.(((((.......))))).)))..)).))))).))..)....)))))) ( -25.40) >DroSec_CAF1 295073 106 - 1 UU---UUUUAUCGUCUUAAAUUUACAAUUCAAUGUCAUGCCGGCUAUAA-----UGUGGUUGCUUCUUCCAUAUUUCCAAAUUGAGGCAAAACGAAGAAGAAGUUGCU------GCUGCA ((---((((.((((.........(((......)))..((((......((-----(((((.........)))))))..........))))..)))).))))))..(((.------...))) ( -16.89) >DroSim_CAF1 316687 109 - 1 UU---UUUUAUCGUCUGAAAUUUACAAUUCAAUGUCAUGCCGGCUAUAU-----UGUGGUUGCUUCUUCCAUAUUGCCAAAUUGAGGCAAAAUGAAGAAGAAGUUGCUGC---UGCUGCA ..---......(((.((((........)))))))...(((.(((.....-----.(..((.(((((((((((.(((((.......))))).)))..)))))))).))..)---.)))))) ( -28.30) >DroYak_CAF1 307357 106 - 1 --------UAUCGCCCGCAAUUUACAAUUCAAUGUCAUGCCGGCUAUAUGGCUAUGUGGCUUCUUCUUUCAUAUUGCCAAAUUGAGGCAAAACGAAGAAGAAGCUGCU------GCUGCA --------....(((.(((....(((......)))..))).))).....(((...(..(((((((((((....(((((.......)))))...)))))))))))..).------)))... ( -36.80) >consensus UU___UUUUAUCGUCUGAAAUUUACAAUUCAAUGUCAUGCCGGCUAUAU_____UGUGGUUGCUUCUUCCAUAUUGCCAAAUUGAGGCAAAACGAAGAAGAAGUUGCU______GCUGCA ........................................((((((((......))))))))(((((((....(((((.......)))))...)))))))..((.((.......)).)). (-19.11 = -19.30 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:31 2006